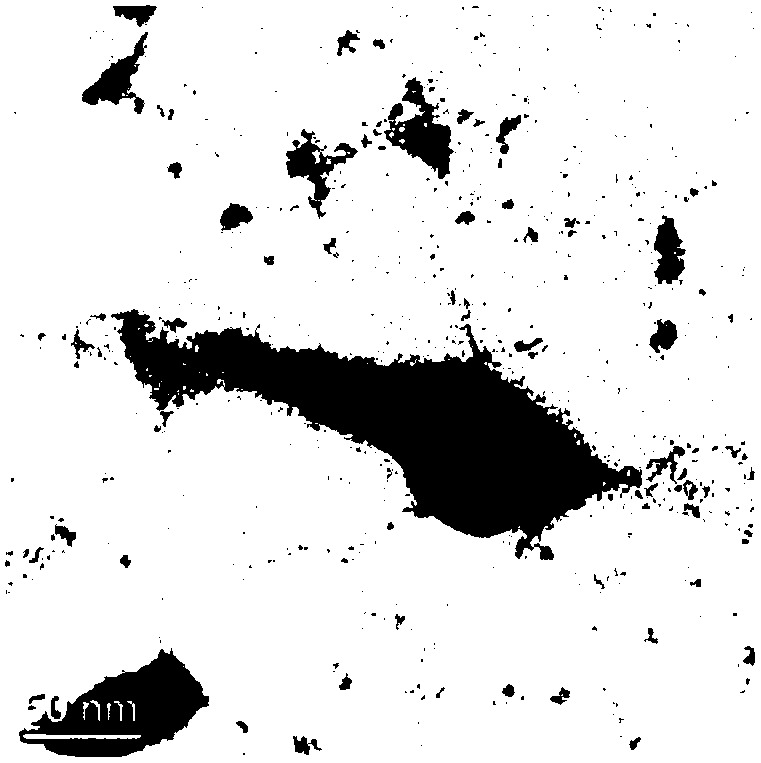
Escherichia coli bacteriophage Esc-COP-7 and pathogenic Escherichia coli proliferation inhibition application thereof
A technology of esc-cop-7 and Escherichia coli, which is applied in the direction of phage, virus/phage, medical raw materials derived from virus/phage, etc., to achieve high specificity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Isolation of phages that can kill Escherichia coli
[0035] When isolating bacteriophages capable of killing Escherichia coli, samples secured from natural environments were used. In addition, Escherichia coli used for phage isolation has been previously isolated by the present inventors and identified as pathogenic Escherichia coli.
[0036] Next, the isolation procedure of the phage will be described in detail. Inoculate the TSB (Tryptic Soy Broth) medium (casein digest, 17g / L; soybean digest, 3g / L; glucose, 2.5g / L; NaCl, 5g / L; Dipotassium hydrogen phosphate, 2.5g / L) was added together with the collected sample (sample), and then cultured with shaking at 37°C for 3-4 hours. After culturing, centrifugation was performed at 8,000 rpm for 20 minutes, and the supernatant was recovered. Escherichia coli was inoculated into the recovered supernatant at a ratio of 1 / 1000, and then cultured again with shaking at 37° C. for 3-4 hours. In the case that the phag...
Embodiment 2
[0041] Embodiment 2: Genome isolation and sequence analysis of phage Esc-COP-7
[0042] The genome of phage Esc-COP-7 was isolated as follows. The genome was isolated using the phage suspension obtained in the same manner as in Example 1. First, in order to remove DNA and RNA of Escherichia coli that may be contained in the suspension, the 200 U of DNase I and RNase A were added to the phage suspension, and then placed at 37° C. for 30 minutes. After standing for 30 minutes, in order to eliminate the activity of DNase I and RNase A, 500 μl of 0.5M ethylenediaminetetraacetic acid (EDTA) was added, and then left still for 10 minutes. Then, it was left still at 65° C. for 10 minutes. Next, to disrupt the outer wall of the phage, add proteinase K After 100 μl, it was reacted at 37° C. for 20 minutes. Then, 500 μl of 10% sodium dodecylsulfate (SDS) was added, and then allowed to react at 65° C. for 1 hour. After reacting for 1 hour, add According to the composition rati...
Embodiment 3
[0046] Embodiment 3: the investigation of the ability of phage Esc-COP-7 to kill pathogenic escherichia coli
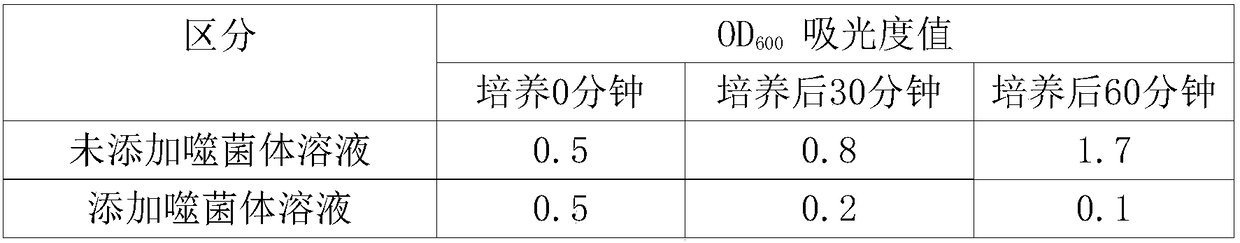
[0047] The ability of the isolated phage Esc-COP-7 to kill pathogenic Escherichia coli was investigated. The killing ability investigation was carried out by investigating whether or not a transparent ring was formed by the spot test proposed in Example 1. The pathogenic Escherichia coli used for the investigation of killing ability was isolated and identified as pathogenic Escherichia coli by the present inventors, and a total of 10 strains were used. Phage Esc-COP-7 has the ability to kill a total of 9 strains of the 10 strains of pathogenic Escherichia coli that became the subject of the experiment. Representative experimental results are as follows: figure 2 shown. In addition, the effect of phage Esc-COP-7 on Bordetella bronchiseptica, Enterococcus faecalis, Enterococcus faecium, Streptococcus mitis, Streptococcus uberis ( Streptococcus uberis) and Pseudomo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com