Method for editing cotton genes
A genome editing, base technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Cloning and optimization of pGhU6-7 promoter and pGhU6-9 promoter
[0050] 1. Blast of the pGhU6 promoter
[0051] Primers were designed using Primer5 software by homologous alignment with the Arabidopsis U6-26 snRNA gene.
[0052] 2. Cloning of the promoter
[0053] Using the genomic DNA of upland cotton YZ1 (Jin et al.2006) as a template, use the following PCR system and program to amplify to obtain two pGhU6, and finally use pGEM-T Easy Vector Systems for cloning and sequencing verification, and separate the promoters with the correct sequence Named as pGhU6-7 promoter, pGhU6-9 promoter. The primer sequences for amplifying the pGhU6-7 promoter are respectively pGhU6-7F (5'-TTAATCTGATGCTCCACCTGCTTTTGAT-3') and pGhU6-7R (5'-CATCTGATGCTTCTTTCGCTTATATAAACG-3'), and the primer sequences for amplifying the pGhU6-9 promoter are respectively pGhU6-9F (5'-TGATTGCTAGAATTTGCAGTAGGCTT-3') and pGhU6-9R (5'-CATCTGATGCTTCTTTCGCTTATAAACG-3'). The PCR system is shown i...
Embodiment 2
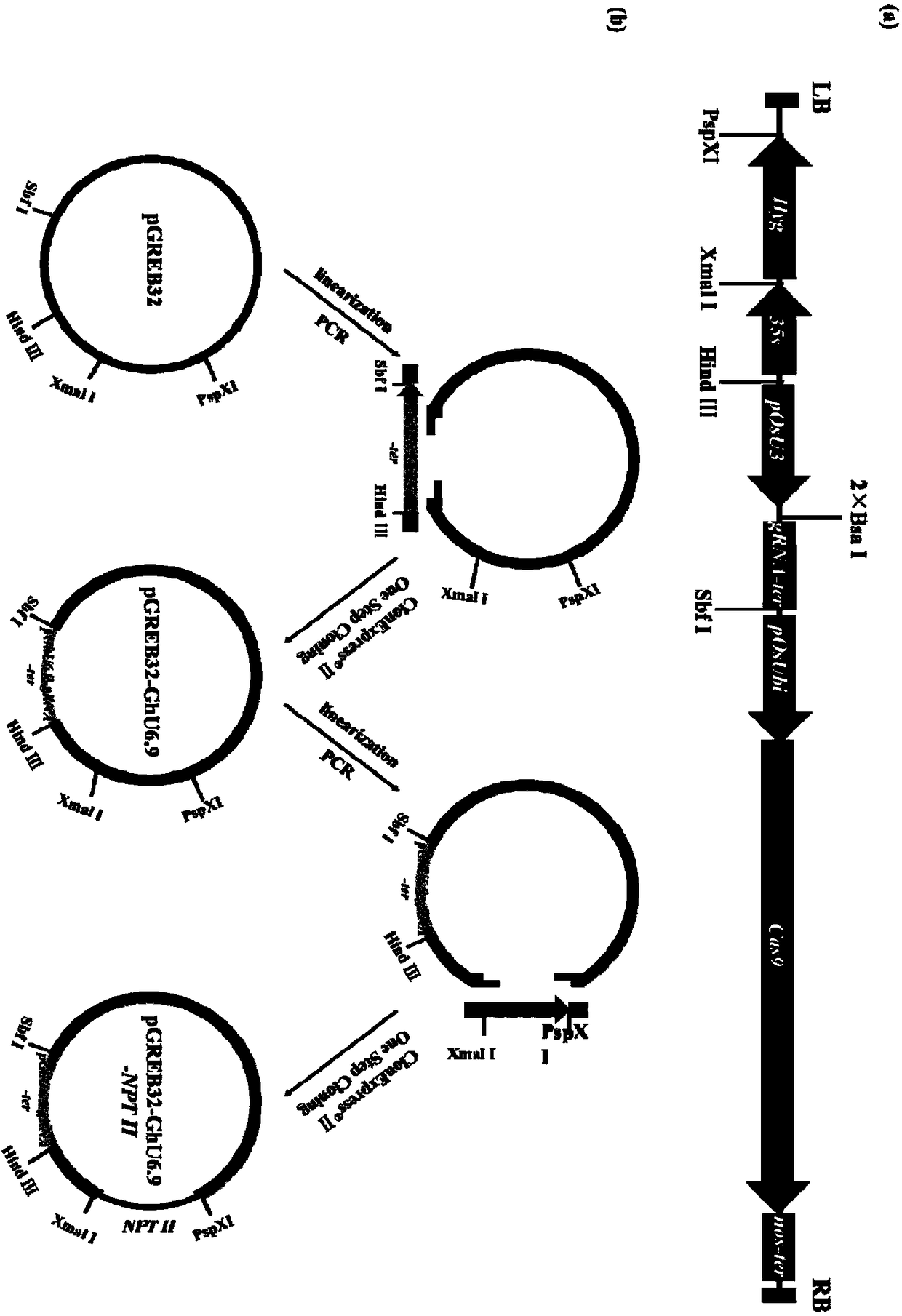
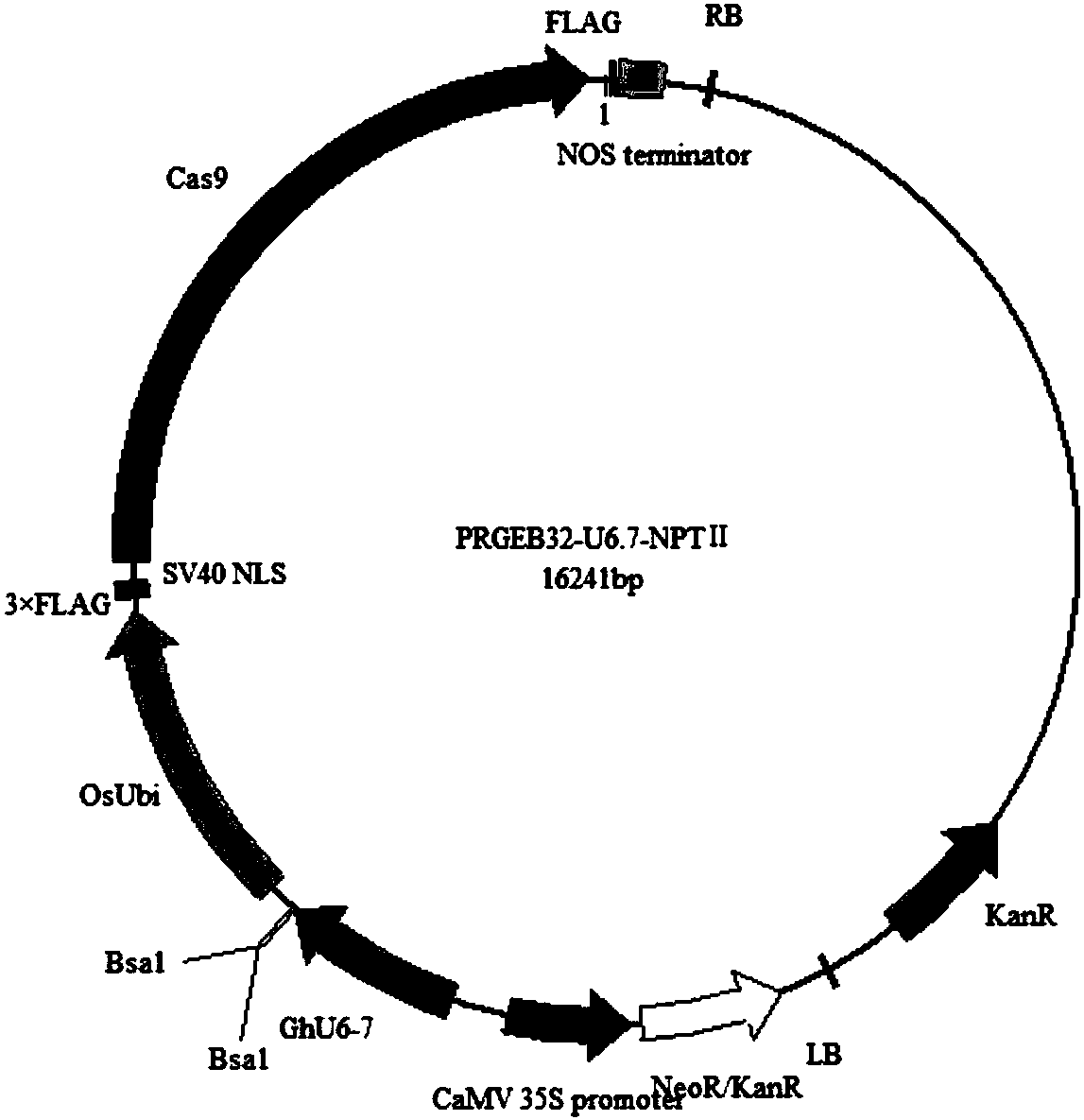
[0060] Example 2: Construction of pRGEB32-GhU6.7-NPTII and pRGEB32-GhU6.9-NPTII vectors
[0061] 1. Connection of pRGEB32 with pGhU6-7I and pGhU6-9I
[0062] Firstly, the pRGEB32 vector was digested, and the digestion system was shown in Table 3, placed at 37°C for 5 hours, and then the digested product was purified using a gel recovery kit (purchased from Wuhan Huaxin Sunshine Biotechnology Co., Ltd.).
[0063] Table 3 50μL enzyme digestion system
[0064]
[0065] Amplify from plasmids with the correct sequences of pGhU6-7I and pGhU6-9I, introduce BSAⅠ and gRNA sequences by overlapping extension PCR, and then use a gel extraction kit to purify the PCR products, digest the PRGEB32 vector with pGhU6-7I, pGhU6 The -9I fragment was ligated by ClonExpress II One Step Cloning Kit (Vazyme C112-02), transformed into Escherichia coli competent, and positive clones were picked for sequencing, and the sequenced correct plasmids were named pRGEB32-GhU6.7, pRGEB32-GhU6. 9.
[0066]...
Embodiment 3
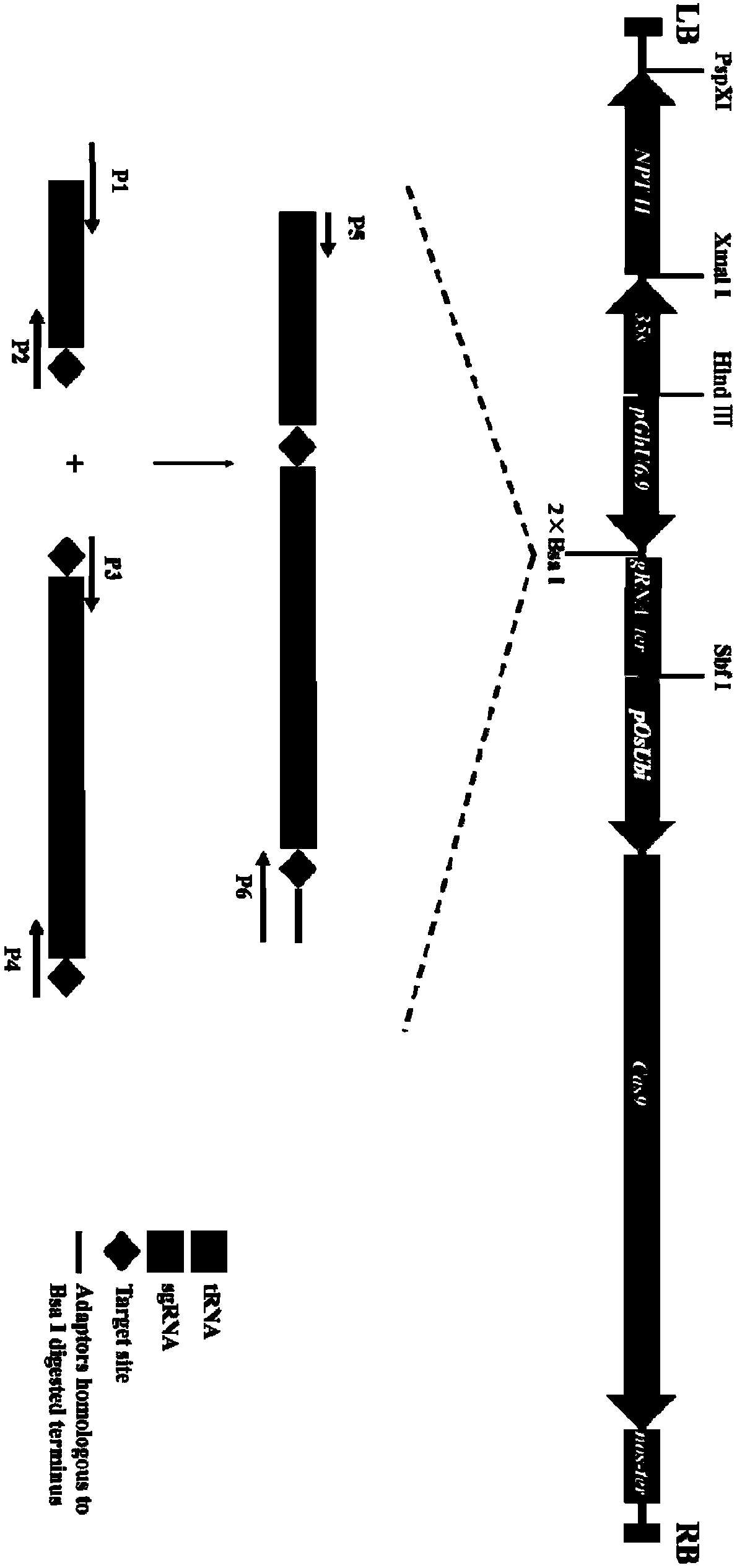
[0077] Example 3: Construction of pRGEB32-GhU6.7-NPTⅡ-sgRNA and pRGEB32-GhU6.9-NPTⅡ-sgRNA vectors
[0078] 1. sgRNA design of GhCLA gene
[0079] Four sgRNAs were designed in the exon region of Gh_A10G2292 gene of upland cotton 1-deoxyxylulose-5-phosphate synthase (Cloroplasto alterados, CLA), named sgRNA1, sgRNA2, sgRNA3, sgRNA4 respectively.
[0080] Table 7GhCLA gene sgRNA statistical table
[0081]
[0082] 2. sgRNA is connected to pRGEB32-GhU6.7-NPTⅡ, pRGEB32-GhU6.9-NPTⅡ vector
[0083]The sequence inserted into the vector is the repeat sequence of tRNA and gRNA, and the repeat sequence is spliced by overlapping extension PCR (Urban et al.1997). The target is added with a primer adapter, and then connected to the vector by a one-step cloning method. Using gRNA2 and gRNA4 as combination 1, and gRNA1 and gRNA3 as combination 2, they were ligated with pRGEB32-GhU6.7-NPTⅡ and pRGEB32-GhU6.9-NPTⅡ vectors respectively. The successfully constructed vectors were named 7NC1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com