Visual quantitative screening new method for dual genes of transgenic crops
A screening and genetic technology, applied in the direction of biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of lack of official positive declaration, lack of scientificity, etc., and achieve the effect of simple and efficient screening and analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Embodiment 1, the establishment of a double colorimetric sensing method for screening the 35s promoter of cauliflower mosaic virus (CaMV) and the terminator of nopaline synthase gene (Nos) by ultra-fast PCR
[0081] (1) Experimental materials
[0082] The nucleotide sequences of the primers designed in this example are shown in Table 1 and the sequence listing.
[0083] Table 1
[0084]
[0085] In Table 1, the upstream primer 35s-F is obtained by carrying out biotin (Biotin) labeling at the 5' end of the nucleotide sequence shown in SEQ ID No. 1 in the sequence listing; the downstream primer 35s-R is obtained by marking the The 3' end of the nucleotide sequence shown in SEQ ID No.: 2 in the sequence listing is obtained after Cy5 labeling; the upstream primer Nos-F is obtained by the nucleotide sequence shown in SEQ ID No.: 3 in the sequence listing The 5' end was labeled with biotin (Biotin); the upstream primer Nos-R was labeled with Digoxin (Digoxin) at the 3' e...
Embodiment 2
[0110] Embodiment 2, sensitivity experiment
[0111] The transgenic crop CBH-351-corn seeds were ground into powder using a ball mill, and the genome was extracted using the plant genome extraction kit of Quanshijin Company, and the amplification reaction was carried out with reference to the double ultra-fast PCR reaction described in step (3) of Example 1 above, wherein The genomes that have been serially diluted are used as templates, and the amount of the templates, that is, the contents of the transgenic components after serial dilution are: (A) 100%, (B) 50%, (C) 5%, (D) 0.5%, ( E) 0.05%, (F) 0%, the percentages are mass percentages.
[0112] After the amplification reaction is completed, mix 10 μL of the dual ultra-fast PCR reaction system with 60 μL of detection buffer (4×SSC, 2% BSA by mass, 0.05% by mass Tween-20, pH 7.0) and mix evenly. Immerse the colloidal gold immunochromatography test paper prepared above, react at room temperature for 5 minutes, and observe th...
Embodiment 3
[0114] Embodiment 3, specificity experiment
[0115]The transgenic crops 59122-corn seeds, GA21-corn seeds, MON809-corn seeds, 32138-corn seeds, 3272-corn seeds, CBH-351-corn seeds, MON87411-corn seeds, 33121-corn seeds are ground into powder using a ball mill, Genomes were extracted using Quanshijin Plant Genome Extraction Kit, and the extracted genomes were used as templates (add 2 uL to each template), and the double ultra-fast PCR reactions were carried out with reference to the double ultra-fast PCR reaction described in step (3) of Example 1 above. .
[0116] After the amplification reaction is completed, mix 10 μL of the dual ultra-fast PCR reaction system with 60 μL of detection buffer (4×SSC, 2% BSA by mass, 0.05% by mass Tween-20, pH 7.0) and mix evenly. Immerse the colloidal gold immunochromatography test paper prepared above, react at room temperature for 5 minutes, and observe the results.
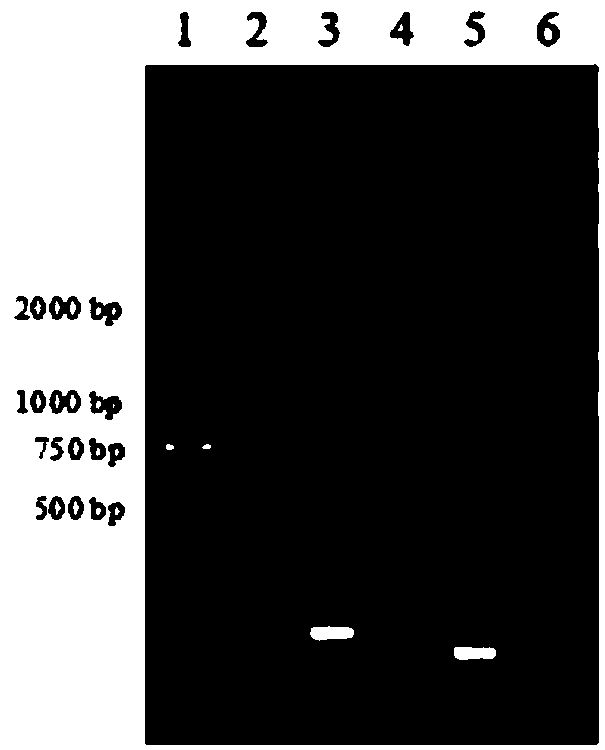
[0117] Experimental results such as Figure 4 and shown in Table 4. T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com