Mulberry-derived enterobacter cloacae specific sequence, primer group Yt4 as well as applications of specific sequence and primer group Yt4 in detection of enterobacter cloacae
A technology of Enterobacter cloacae and specific sequence, applied in the directions of microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the difficulty, low sensitivity and low efficiency of the detection and identification of mulberry wilt. and other problems, to achieve the effect of reliable detection results, high sensitivity and convenient use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Obtaining the specific gene sequence of Enterobacter cloacae
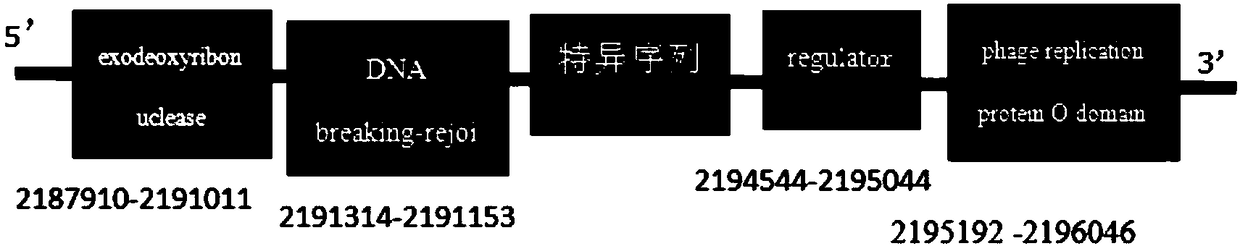
[0064] Using the high-throughput whole-genome sequencing method, the whole genome sequence of Enterobacter cloacae was assembled, and a total of 4.73Mb whole genome sequence was assembled. The number of G bases was 1039163 bp, accounting for 28.04%; the number of C bases was 1325068 bp, accounting for The ratio is 28.01%, and the ratio of GC is 56.05%, encoding 4600 genes; the number of A bases is 1039163 bp, accounting for 21.95%; the number of T bases is 1325068 bp, accounting for 28%. Enterobacter cloacae was found in the soil of the branches and roots of diseased mulberries. The Enterobacter cloacae from the mulberry source was significantly different from other Enterobacter cloacae, and the similarity to the strain with the highest similarity (AP018340.1) was 97%. through
[0065] Based on the comparative analysis of the whole genome of Enterobacter cloacae, the specific gene sequence was obtained...
Embodiment 2
[0066] Example 2 Construction and sequencing analysis of specific gene cloning plasmid based on Enterobacter mulberry
[0067] The DNA of Enterobacter cloacae was used as the PCR amplification template, and the primer 5f / 2007r (5f: ggtgtctggacaatctcagt; 2007r: catttcaaccagttacgata) was used for PCR amplification.
[0068] After PCR amplification products are detected by agarose gel electrophoresis, the specific target fragments are recovered under ultraviolet light, and the target fragments are recovered according to the steps of the DNA rapid recovery / purification kit (purchased from Beijing Dingguo Biotechnology Co., Ltd.).
[0069] Vector connection: The purpose of recovery is to connect with pEASY@-Blunt vector (system: PCR product 1 µL, pEASY@-Blunt vector 1 µL, dH2O 3 µL), mix well and ligate at 25°C for 15 min.
[0070] Transformation of the ligation product: add the ligation product and 50 µL of newly thawed Trans1-T1 competent cells, mix gently, ice bath for 25 minutes, activa...
Embodiment 3
[0073] Example 3 Design of primers for detection of Enterobacter cloacae and establishment of LAMP amplification method
[0074] 1. Primer design and screening
[0075] Based on the specific gene of Enterobacter cloacae obtained in Example 1 (shown in SEQ ID NO.1), a large number of primers were designed, and after analysis and screening, a set of primer sets with excellent specificity and sensitivity were finally obtained , Named Yt4. The sequence of the primer set is shown in Table 1 below:
[0076] Table 1 The sequence of primer set Yt4
[0077]
[0078] 2. Establishment of LAMP amplification method
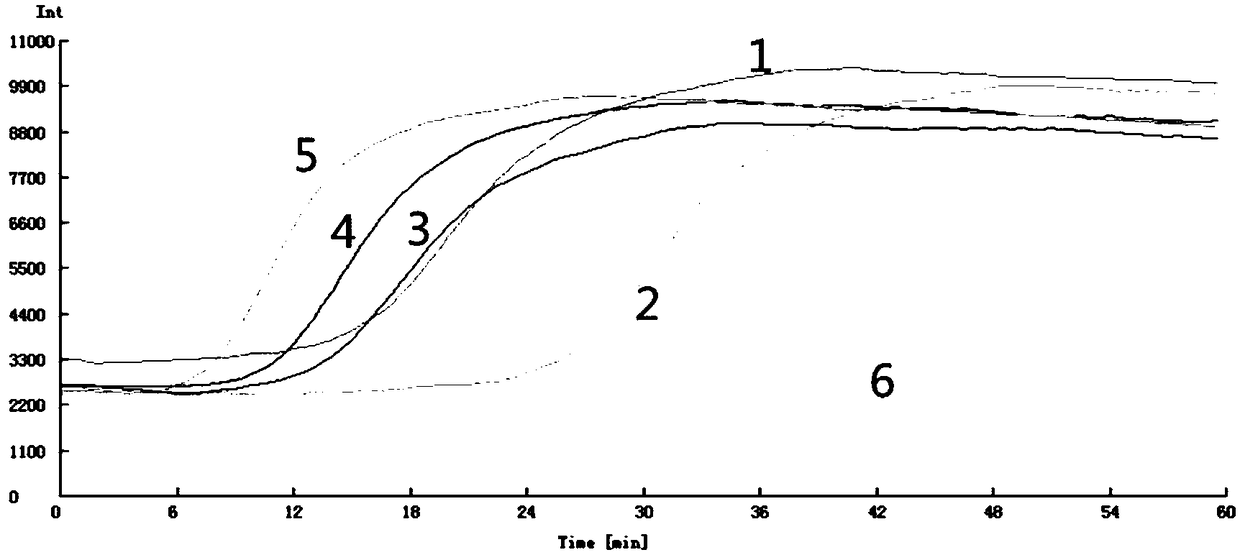
[0079] (1) Optimization of LAMP amplification reaction temperature
[0080] Using 5 ng / μL of Enterobacter cloacae DNA as template, and Yt4 primer set as amplification primers, at 60℃, 61℃, 62℃, 63℃, 64℃, 65℃ and other 6 different reaction temperatures Perform LAMP amplification with 25μL amplification system (as shown in Table 3).
[0081] (2) Optimization of the concentration ratio of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com