Preparation method and application of a CD132 gene-deleted immunodeficiency animal model
A technology of immunodeficiency and animal models, applied in the field of animal genetic engineering, can solve the problems of affecting experimental results, impure background, long cycle, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0168] Example 1 Design of CD132 gene sgRNA
[0169] The target sequence determines the targeting specificity of the sgRNA and the efficiency of inducing Cas9 to cleave the target gene. Therefore, efficient and specific target sequence selection and design are the prerequisites for constructing sgRNA expression vectors.
[0170] Multiple sgRNAs are designed for mouse immune-related genes, where the target site sequence targeted by each sgRNA is as follows:
[0171] sgRNA-1 target site sequence (SEQ ID NO: 1): 5'-ccaccggaagctacgacaaaagg-3'
[0172] sgRNA-2 target site sequence (SEQ ID NO: 2): 5'-tctctacagcgtggtttctaagg-3'
[0173] sgRNA-3 target site sequence (SEQ ID NO: 3): 5'-ggcttgtgggagagtggttcagg-3'
[0174] sgRNA-4 target site sequence (SEQ ID NO: 4): 5'-ccacgctgtagagagagggggggg-3'
[0175] sgRNA-5 target site sequence (SEQ ID NO: 5): 5'-agggggaggttagcgtcacttagg-3'
[0176] sgRNA-6 target site sequence (SEQ ID NO: 6): 5'-gaaatcgaaacttagccccaagg-3'
[0177] sgRNA-7 t...
Embodiment 2
[0181] Example 2 Screening of CD132 gene sgRNA
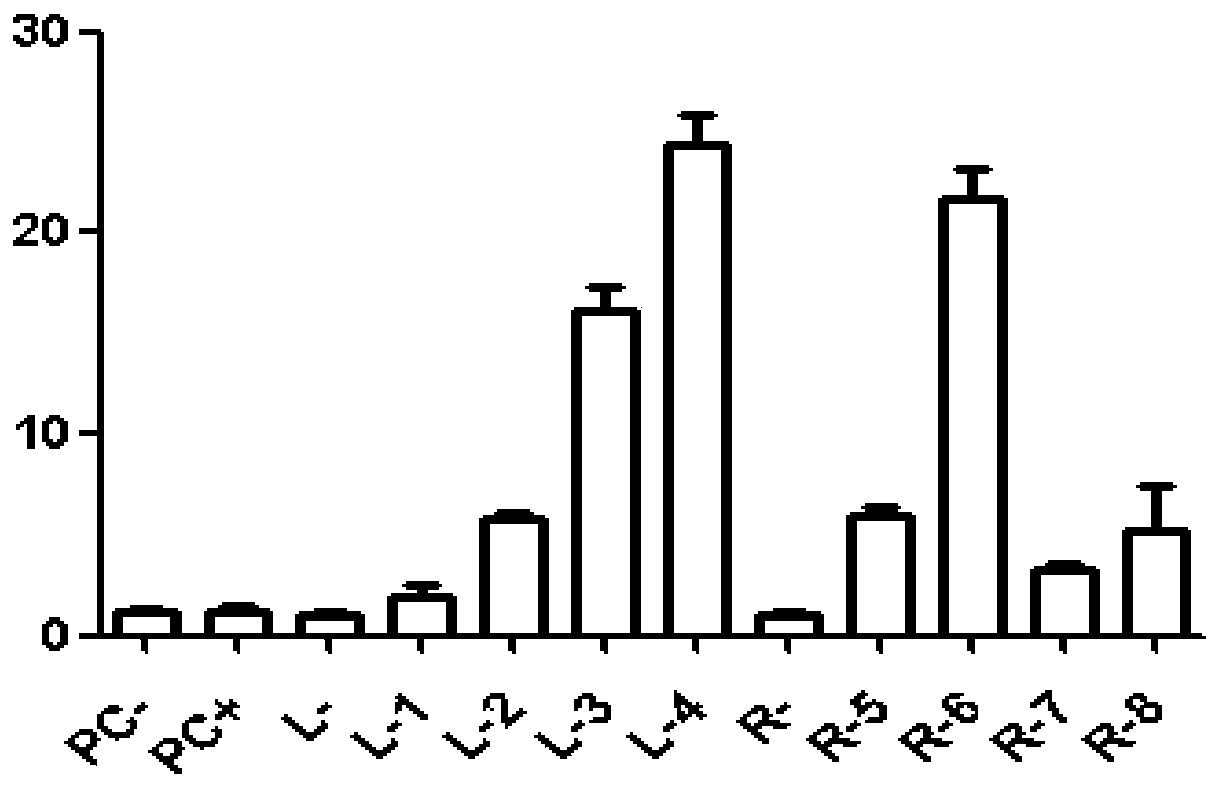
[0182] UCA kit was used to detect the activities of multiple sgRNAs. The results showed that the sgRNAs had different activities. For the test results, see figure 1 . Among them, the activities of sgRNA-1 and sgRNA-7 are relatively low, which may be caused by the particularity of the target site sequence, but according to our experiments, the values of L-1 and R-7 are still significantly higher than those of the control group, and can still be Judging that sgRNA-1 and sgRNA-7 are active, the activity meets the requirements of gene targeting experiments. According to the activity detection results, sgRNA3 (relative activity value of 16.1±1.2) and sgRNA6 (relative activity value of 21.7±1.5) were preferred for subsequent experiments. Remove the three NGG bases at the 3' end of the sgRNA sequence to obtain the upper and lower single strands as follows:
[0183] sgRNA3: upstream: 5'-ggcttgtgggagagtggttc-3' (SEQ ID NO: 18)
[0...
Embodiment 3
[0190] Example 3 pT7sgRNA-G2 plasmid construction
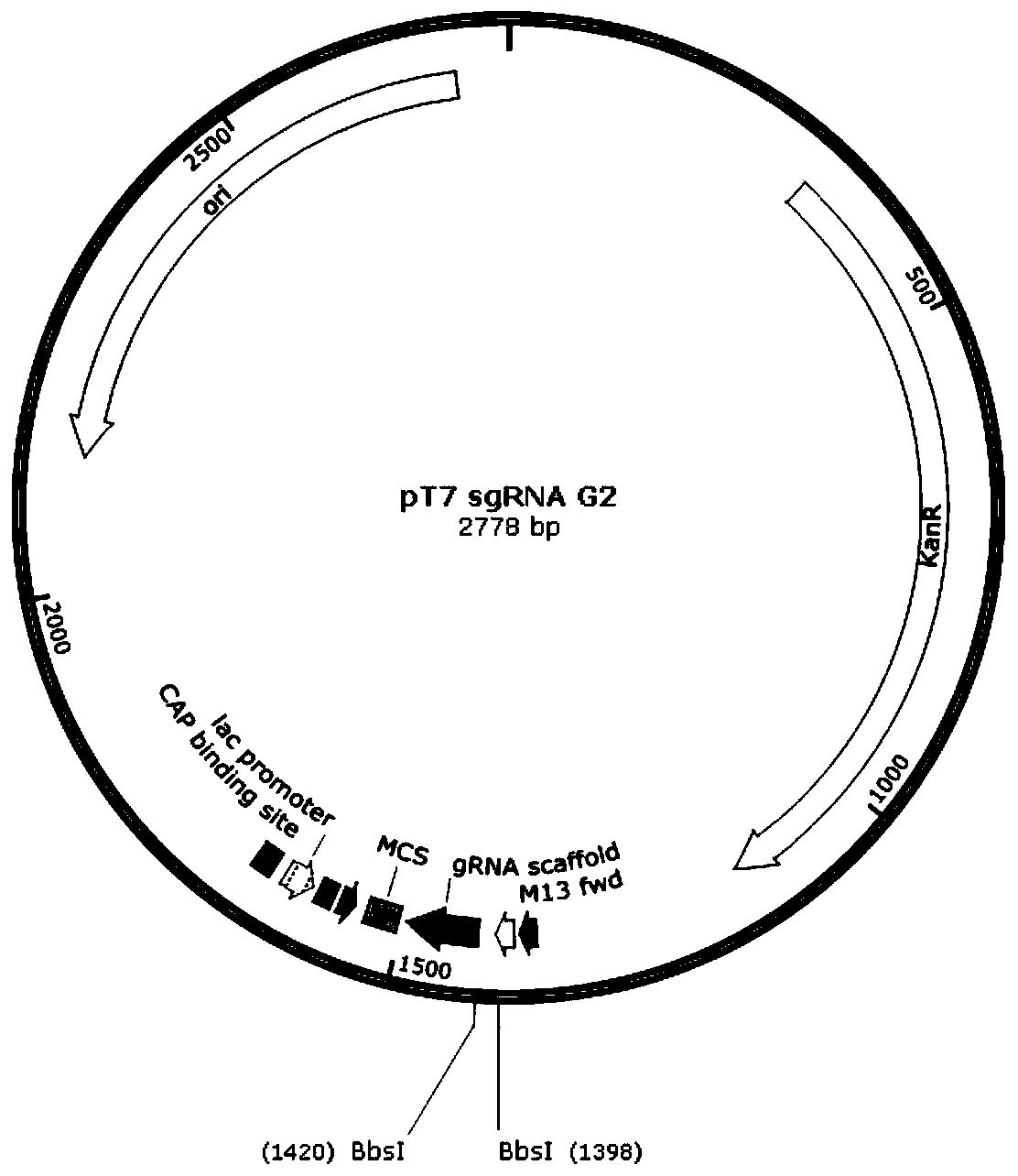
[0191] The fragmented DNA containing the T7 promoter and sgRNA scaffold was synthesized by a plasmid synthesis company and ligated to the backbone vector pHSG299 by restriction enzyme digestion (EcoRI and BamHI) in sequence. After sequencing verification by a professional sequencing company, the results showed that the target plasmid: pT7sgRNA-G2 plasmid was obtained , see the plasmid map of pT7sgRNA-G2 figure 2 .
[0192] Fragment DNA containing T7 promoter and sgRNA scaffold (SEQ ID NO: 13):
[0193] gaattctaatacgactcactataggggtcttcgagaagacctgttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgcttttaaaggatcc
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com