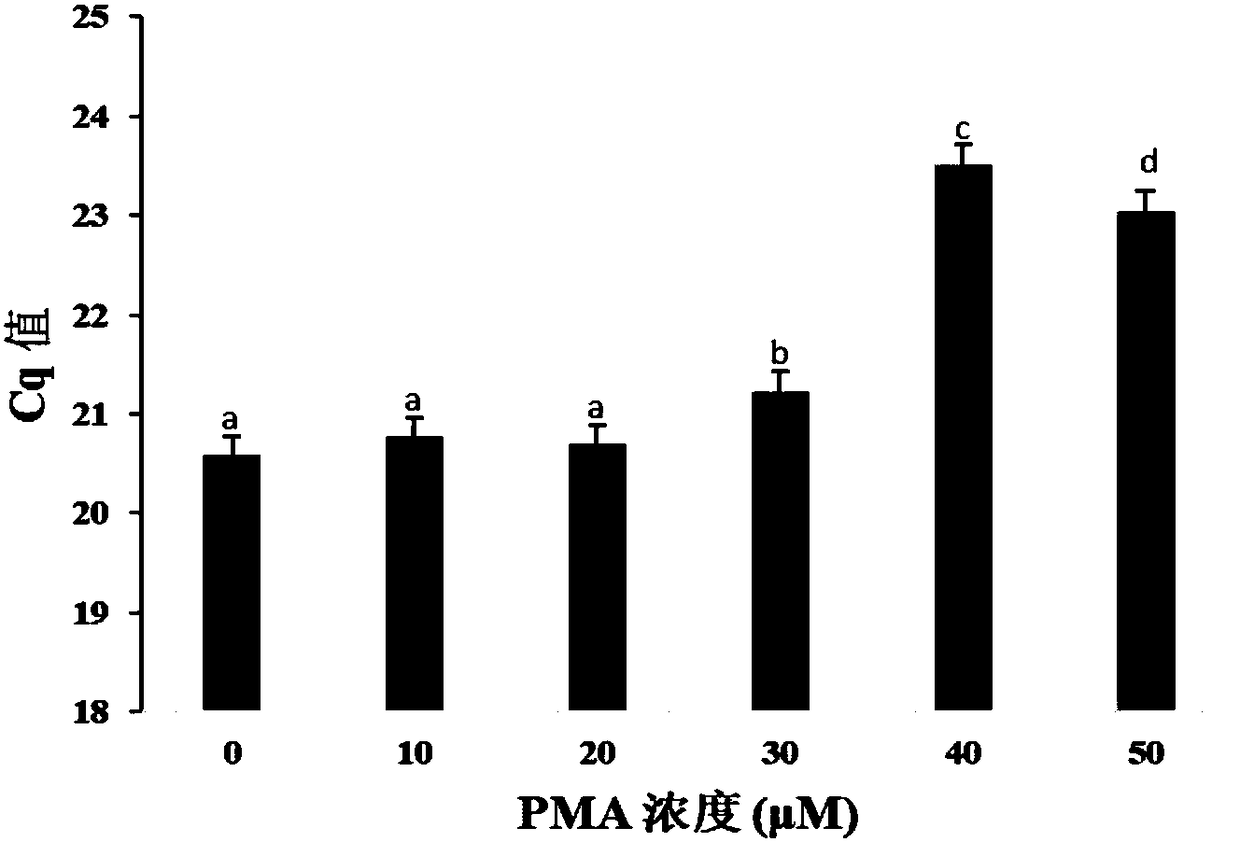
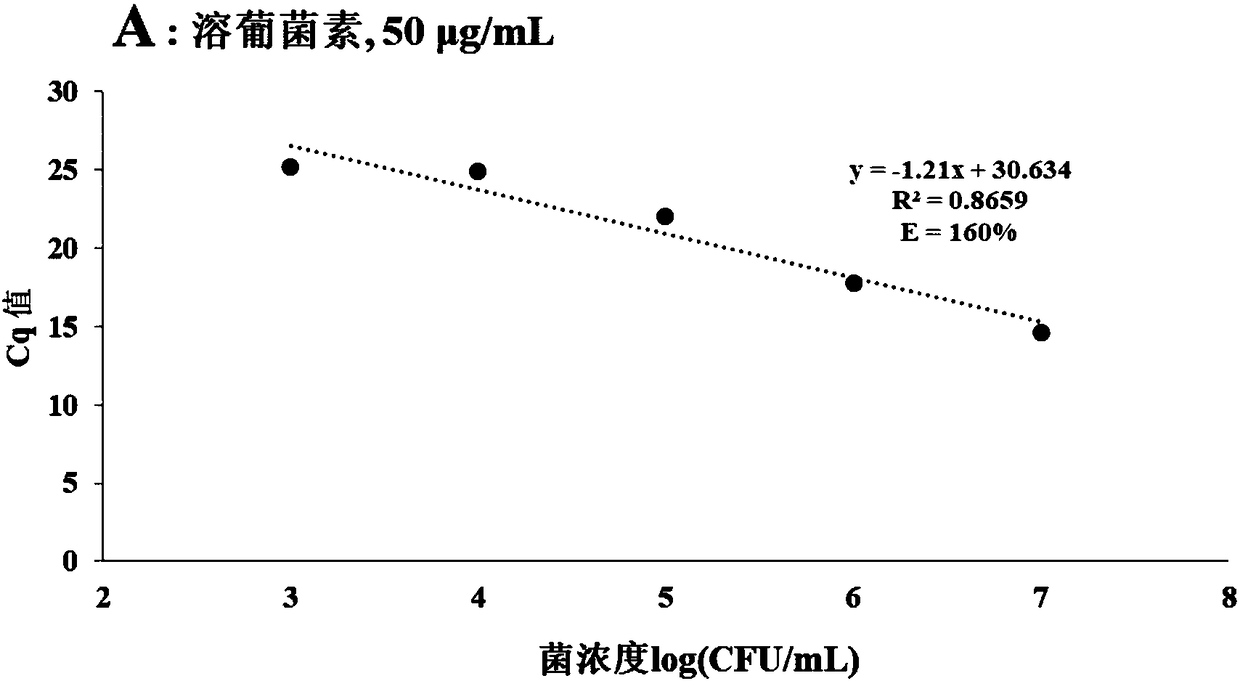
Method for detecting viable bacteria in staphylococcus aureus
A staphylococcus and lysostaphin technology, applied in the field of microbial detection, can solve problems such as inability to effectively distinguish live bacteria from dead bacteria, high detection values, and inaccurate detection results, so as to avoid false positive results and achieve high extraction efficiency , the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0054] 3. Preparation of dead bacteria
[0055] Centrifuge the bacterial suspension at 5000 g for 10 min at 4°C, discard the supernatant, and resuspend the bacterial cells with 0.1% peptone water. The concentration of the liquid after resuspension was adjusted to 10 7 -10 8 CFU / mL, and divided into two equally. One of them was heated at 90°C for 20 minutes to prepare dead bacteria, and the other was used as live bacteria.
Embodiment 1
[0057] Primer inclusion and exclusivity verification
[0058] Using 18 strains, including 10 standard strains of Staphylococcus aureus and 8 other known strains, bacterial DNA was extracted by CTAB method. The PCR reaction system (25 μL) included 12.5 μL Master Mix, 2 μL template DNA, 1 μL each of upstream and downstream primers, the concentration of the primers was 4000 nM, and 8.5 μL of water.
[0059] The primer sequence is: upstream primer 5'-CACCTGAAACAAAGCATCCTAAA-3', downstream primer 5'-CGCTAAGCCACGTCCATATT-3';
[0060] Reaction program: pre-denaturation at 95°C for 10 min, denaturation at 95°C for 15 s, annealing at 62°C for 1 min, and 40 cycles. The amplified products were subjected to electrophoresis in 2.5% agarose gel electrophoresis prepared in 1×TE buffer.
[0061] As shown in Table 1, using the above primers, only the DNA of Staphylococcus aureus showed positive signals, and the DNA of other strains showed no positive signals. This result indicated that the ...
Embodiment 2
[0065] Optimization of SDS concentration
[0066] SDS was dissolved in 0.1% peptone water to prepare a 20% SDS stock solution, which was then sterilized.
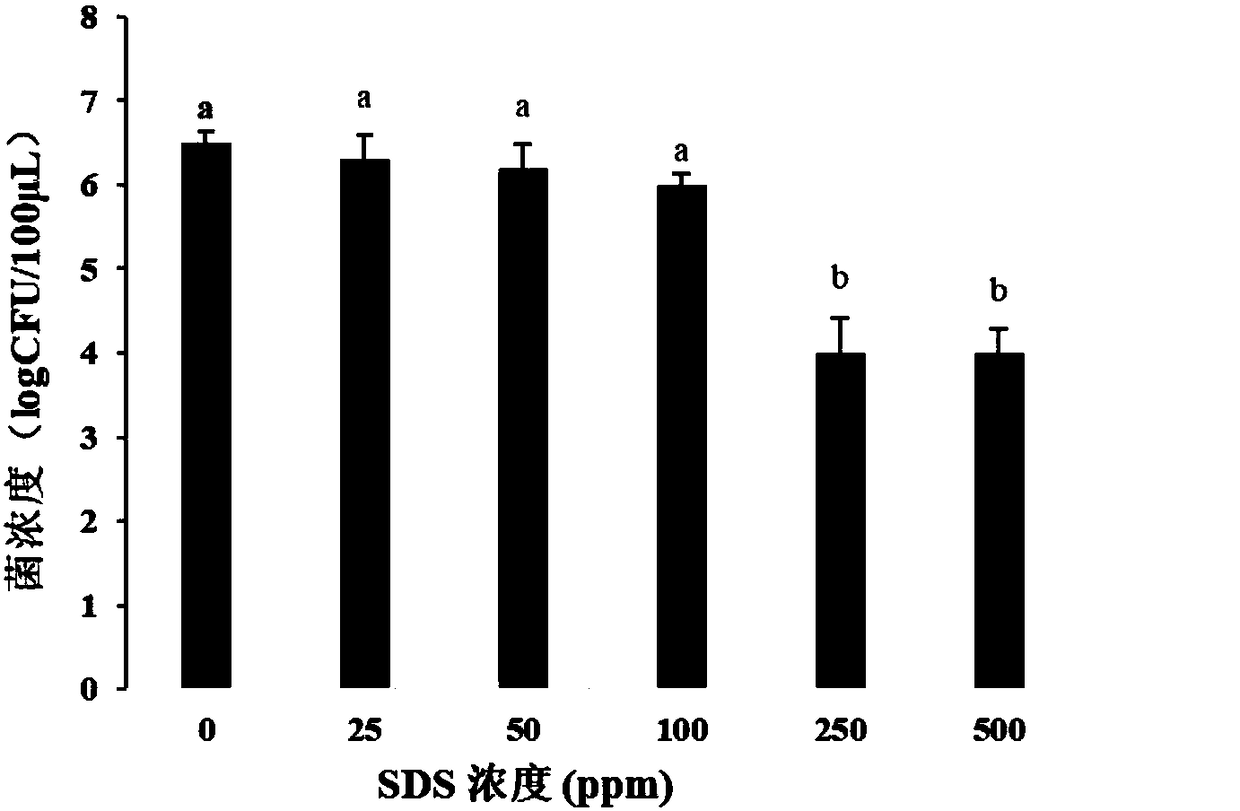
[0067] Staphylococcus aureus (ATCC6538) bacteria liquid (10 7 CFU / mL) was heat-treated at 90°C for 35s, and centrifuged at 5000g for 10min at 4°C. Then resuspended with different concentrations of SDS, the concentrations of SDS were 0, 25, 50, 100, 250, 500, 1000ppm. Take 100 μL of the solution and spread it on a plate, incubate at 37°C for 24 hours, and observe the number of colonies. Select the optimal concentration according to the inhibition of SDS to Staphylococcus aureus live bacteria.
[0068] Such as figure 1 As shown, when the SDS concentration is 0, 25, 50, and 100 ppm, the corresponding logCFU values are 6.5, 6.3, 6.2, and 6.0, respectively. When the SDS concentration was 250, 500, 1000ppm, the logCFU values were 4, 4, 0, respectively. It can be seen from the figure that when the SDS concentration is 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com