Method for detecting rhinovirus typing and enterovirus
A RT-PCR, specific technology, applied in the detection of rhinovirus typing and enterovirus, identification of HRVA, HRVB, EV, HRVC field, can solve the problems of complex operation, high mutation rate, indistinguishable, etc., to achieve throughput High, short time, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1: Collection of Samples and Extraction of Nucleic Acids
[0066] 1. Sample collection:
[0067] 1. Sample type: throat swab or nasopharyngeal aspiration.
[0068] 2. Sampling method of throat swab:
[0069] (1) Collection equipment: use special sterile throat swabs (flocked nylon swabs are recommended, cotton swabs and wooden poles are not recommended).
[0070] (2) Sampling solution: Use a sampling solution containing protein stabilizers and antibiotics (bovine serum 5%, penicillin 200U / mL, streptomycin 200U / mL, nystatin 25U / mL), with 2% NaHCO 3 Adjust the pH value to 7.4 (or use isotonic phosphate buffer or normal saline if the above sampling solution is not available)
[0071] (3) Sampling tube: screw cap, frozen at -70°C.
[0072] (4) Sampling method: press the patient's tongue with the tongue depressor with the left hand, extend the swab to the pharynx with the right hand, wipe the posterior pharyngeal wall and tonsils on both sides several times with ...
Embodiment 2
[0079] Embodiment 2: Detection of HRV typing and EV
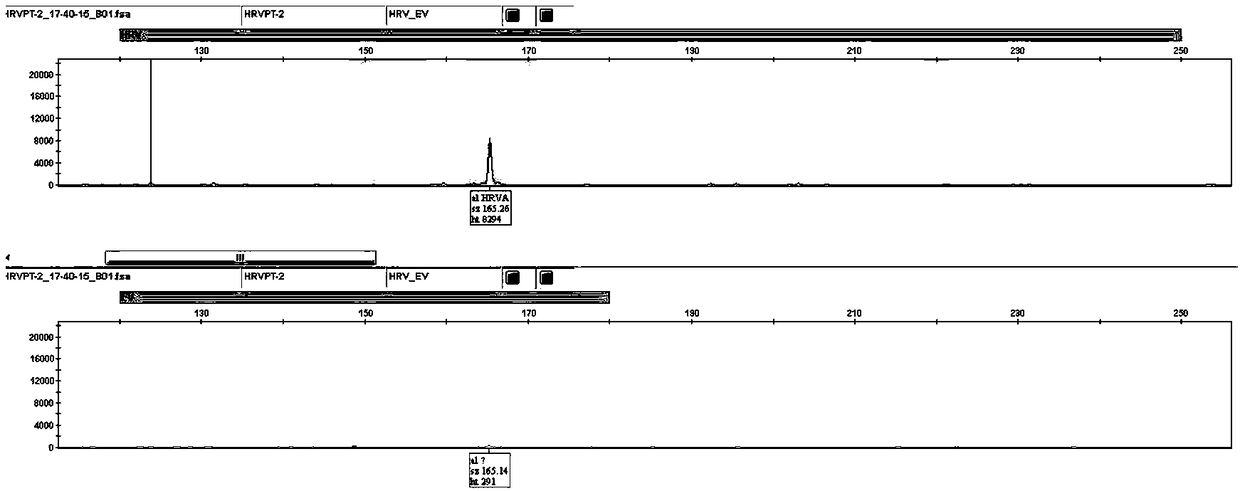
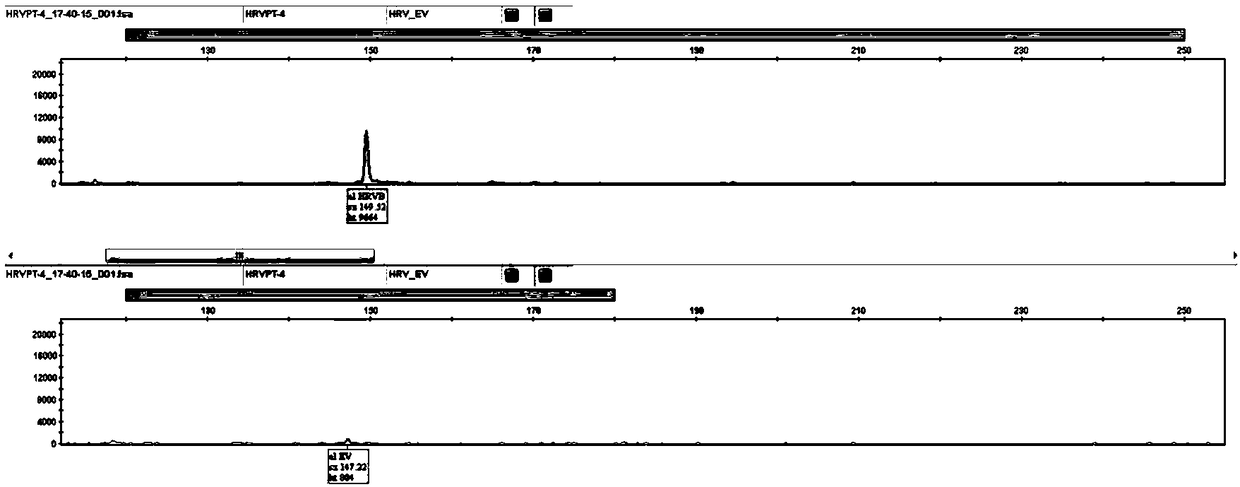
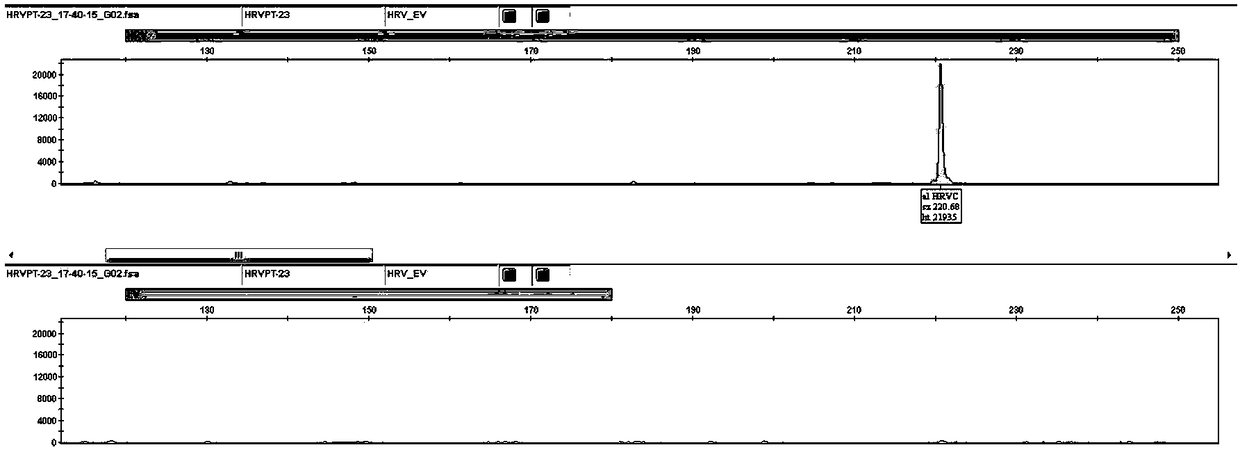
[0080] Samples C7659, B8460, C3604, and C5038 were selected. These four samples corresponded to HRVA, HRVB, HRVC, and EV-positive clinical samples (by virus isolation, culture, and sequencing identification). Throat swabs of patients with respiratory tract infection obtained by the hospital according to the sample collection method described in Example 1. Nucleic acid extraction was performed according to the nucleic acid extraction method in Example 1, and the extracted nucleic acid was used as a template for RT-PCR and electrophoresis analysis.
[0081] Specific steps are as follows:
[0082] 1. Sample pretreatment
[0083] Place the sampling tube on a vortex mixer and vortex fully for 10 seconds to wash off the virus and virus-containing cells attached to the swab, and absorb the corresponding volume of Samples are extracted.
[0084] 2. Nucleic acid extraction
[0085] Use "Nucleic Acid Extraction or Purification R...
Embodiment 3
[0112] Example 3: Verify the identified HRV typing and EV by sequencing
[0113] The nucleic acid extracted from the samples C7659, B8460, C3604, and C5038 in Example 2 was verified by sequencing.
[0114] 1. RT-PCR amplification
[0115] 1.1 Configure RT-PCR system
[0116] 1.1.1 In order to verify the detection accuracy of this method, the inventors used species conservative primers (primers designed based on the IRES region) for PCR amplification and sequencing.
[0117] 1.1.2 Thoroughly vortex the reaction master mix (including sequencing primers, PCR buffer, dNTP) and centrifuge for 10 seconds; Place upright on ice.
[0118] 1.1.3 Add premixed RT-PCR reagents (including the reaction master mix and RT-PCR enzyme solution in 1.1.2) to a 1.5mL centrifuge tube: 28 μL × (N+1) reaction master mix and 2 μL × (N +1) RT-PCR enzyme solution (N is the number of samples, 4 in this embodiment); the mixed solution in the centrifuge tube was inverted and centrifuged briefly, then pl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com