Selection method of reference genes in quantitative real-time PCR analysis of jerusalem artichoke
An internal reference gene, real-time quantitative technology, applied in the field of quantitative PCR
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
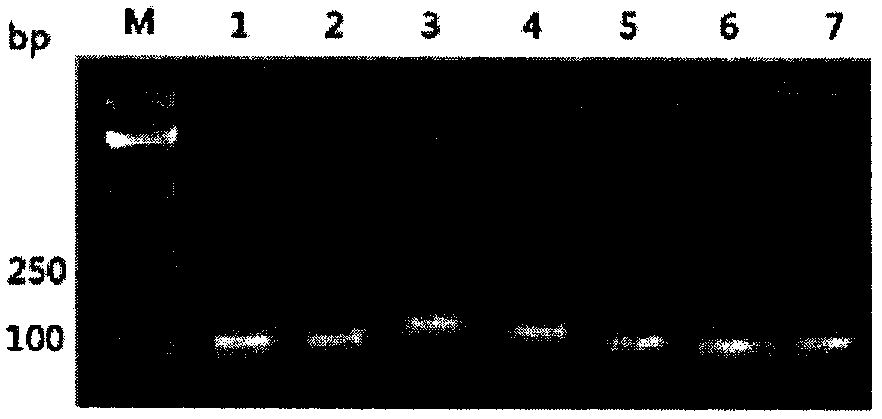
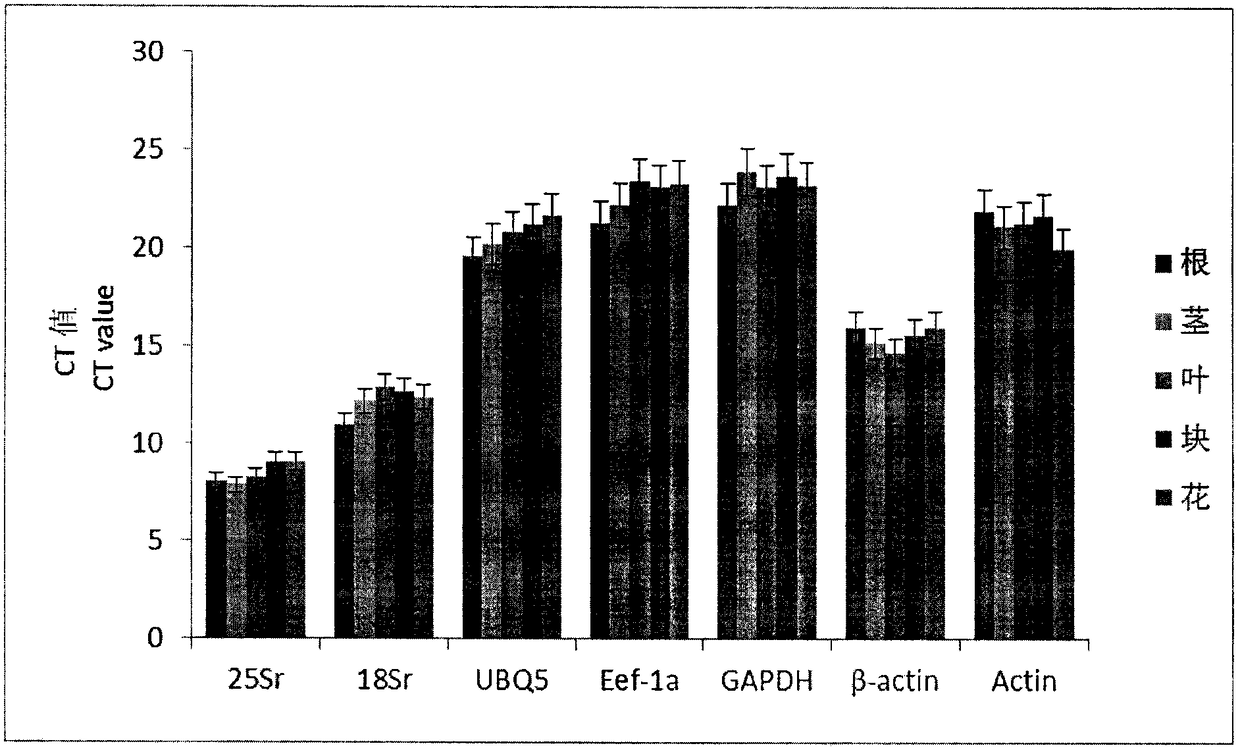
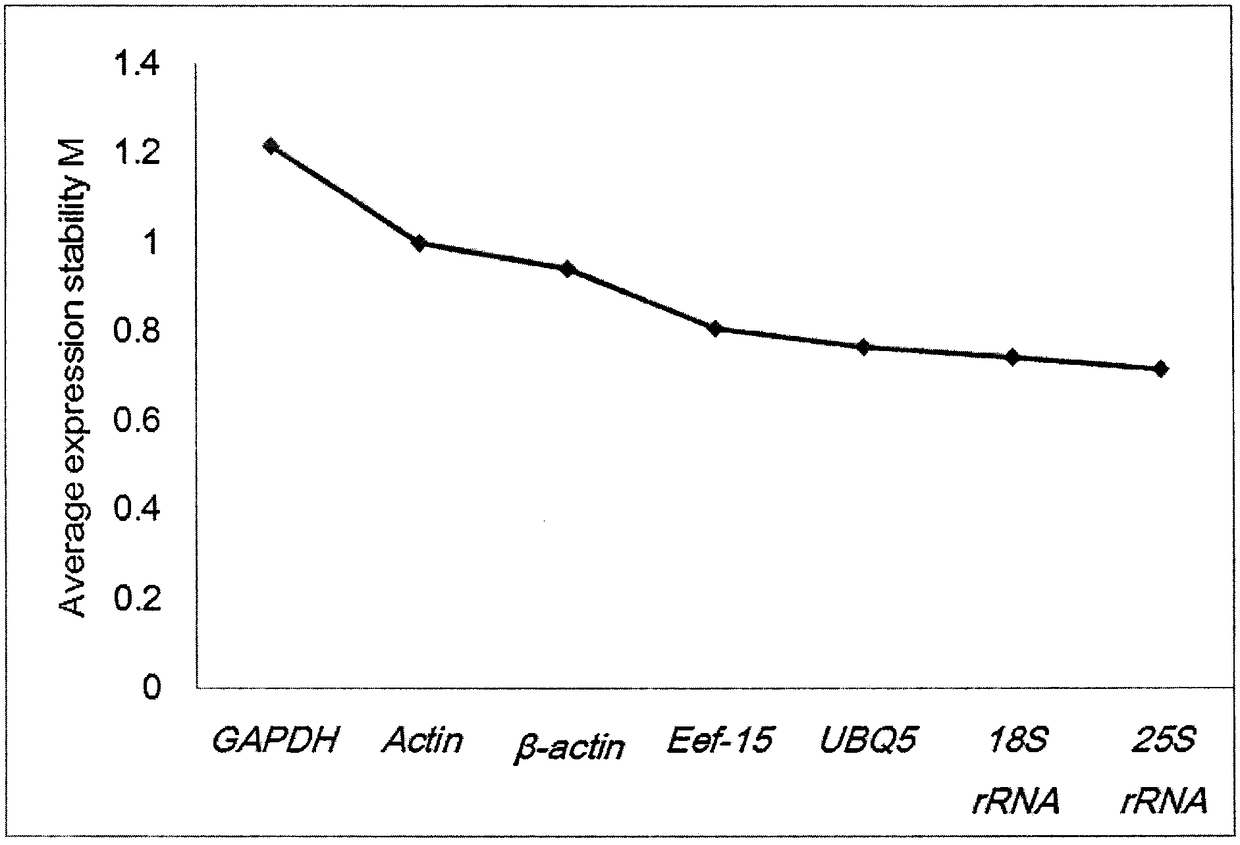
[0016] combine figure 1 , figure 2 and image 3 Show the specific embodiment of the selection method of internal reference gene in a kind of Jerusalem artichoke real-time quantitative PCR analysis of the present invention: The steps of the selection method of internal reference gene in this Jerusalem artichoke real-time quantitative PCR analysis are as follows:
[0017] (1) Extraction of total RNA: Weigh 100mg of Jerusalem artichoke leaf or tuber tissue into a pre-cooled mortar, quickly grind it into powder in liquid nitrogen, and transfer it to a 1.5mL enzyme-free centrifuge tube filled with 1mL Trizol , place the homogenized sample at room temperature for 5min, add 200ul chloroform, cover the tube cap, shake vigorously on a vortex oscillator for about 15sec, then let it stand at room temperature for 3min, 12000r·min -1 Centrifuge at 4°C for 10 minutes, the sample will be divided into three layers, transfer the upper transparent aqueous phase (500ul) to a new enzyme-free c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com