Method and detection kit used for nasal cavity flora detection
A technology of kits and colonies, applied in biochemical equipment and methods, microbial measurement/testing, chemical libraries, etc., can solve the problems that are not conducive to the automatic library construction of large-scale samples and the cumbersome steps of sequencing library construction, and achieve high efficiency Reaction system and method, increased amplification sensitivity, good conservation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1. Preparation and application of kits for detection of nasal flora
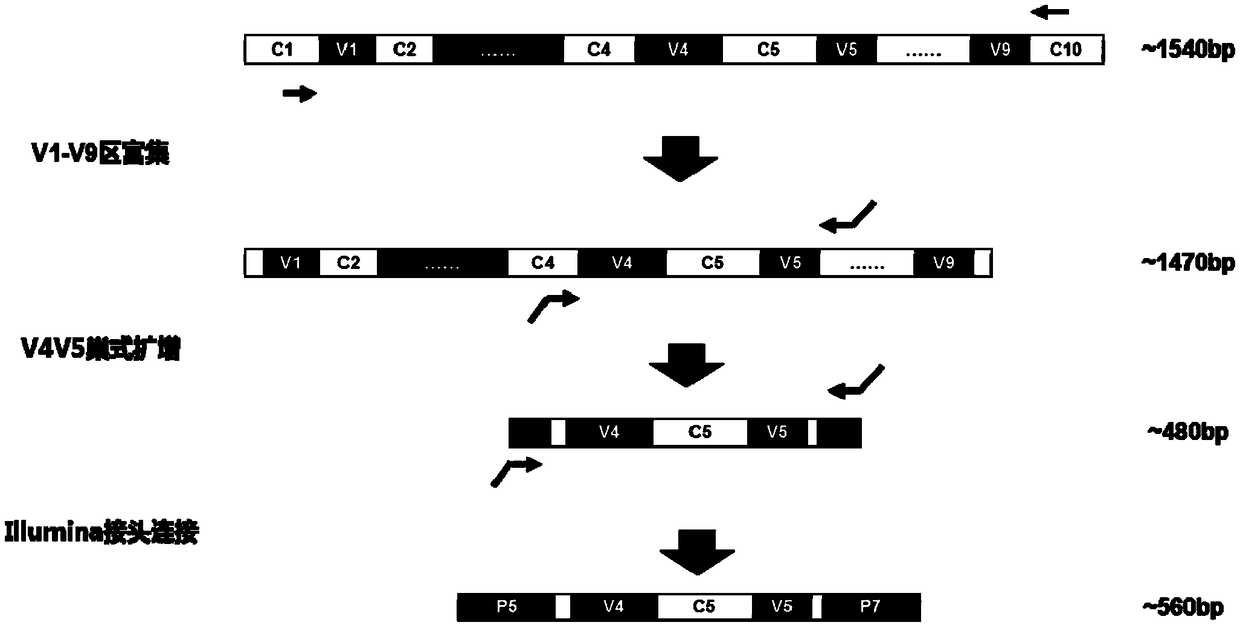
[0062] 1. Design of primers and preparation of detection kits
[0063] 1. Primers for amplifying the V1-V9 region of bacterial 16S rDNA
[0064] Design the following three primers according to the V1-V9 region of bacterial 16S rDNA:
[0065] Upstream primer FP1: sequence 1 in the sequence listing: 5'-AGRGTTYGATYMTGGCTCAG-3' (sequence 1)
[0066] Downstream primer RP1-1: sequence 2 in the sequence listing: 5'-TACAGYTACCWTGTTACGACTTNN-3' (sequence 2)
[0067] Downstream primer RP2-1: sequence 3 in the sequence listing: 5'-GGYTACCTTGTDACGACTTNN-3' (sequence 3)
[0068] 2. Primers for amplifying the V4-V5 region of bacterial 16S rDNA
[0069] Due to the single sequencing product, when performing sequencing on the Illumina platform, the 5-terminus of the PCR product is the same sequence, resulting in a single sequencing start signal, resulting in a serious decline in sequencing quality. The pr...
Embodiment 16
[0160] For the results obtained from the 16S rDNA sequencing of this embodiment, the operational taxonomic unit (OTU) is obtained by using the QIIME software to classify the operational unit (OTU). According to the Greengene database, the representative sequence of the OTU is classified, and the microbial kingdom, phylum, class, order, family, and genus in the sample are obtained. Distribution.
[0161] Made according to the obtained results image 3 The heatmap of the percentage of nasal genus and Figure 4 Histogram of nasal bacterial genera abundance. according to image 3 It can be clearly observed that different individuals and the difference of the left and right nasal bacteria in the same individual. according to Figure 4You can clearly see the proportion of the top 20 dominant bacterial genera in the nasal cavity of each individual, including those mentioned in the published review (Diana M. Proctor and David A. Relma, Cell Host & Microbe, 2017; IF=17.9) There ar...
Embodiment 2
[0162] Embodiment 2, comparison of nested amplification and conventional amplification library
[0163] In order to verify the effect of the nested amplification method on the microbial amplification efficiency in nasal gDNA samples, 9 samples of nasal gDNA samples with Qubit quantitative concentration between 10-100 ng / μL were selected, 10 ng of each sample was mixed and divided into two parts For subsequent database construction.
[0164] Experimental group: The above 2 mixed nasal cavity gDNA samples were constructed according to the sequencing library construction method of 2-3 in Example 1, including 3-step PCR amplification;
[0165] Control group: Different from the experimental group, the enrichment PCR amplification of the V1-V9 region in the first step is not performed, and the enrichment PCR amplification of the V4-V5 region in the second step is directly carried out, as follows: mix the above two parts Nasal cavity gDNA samples were constructed according to the se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com