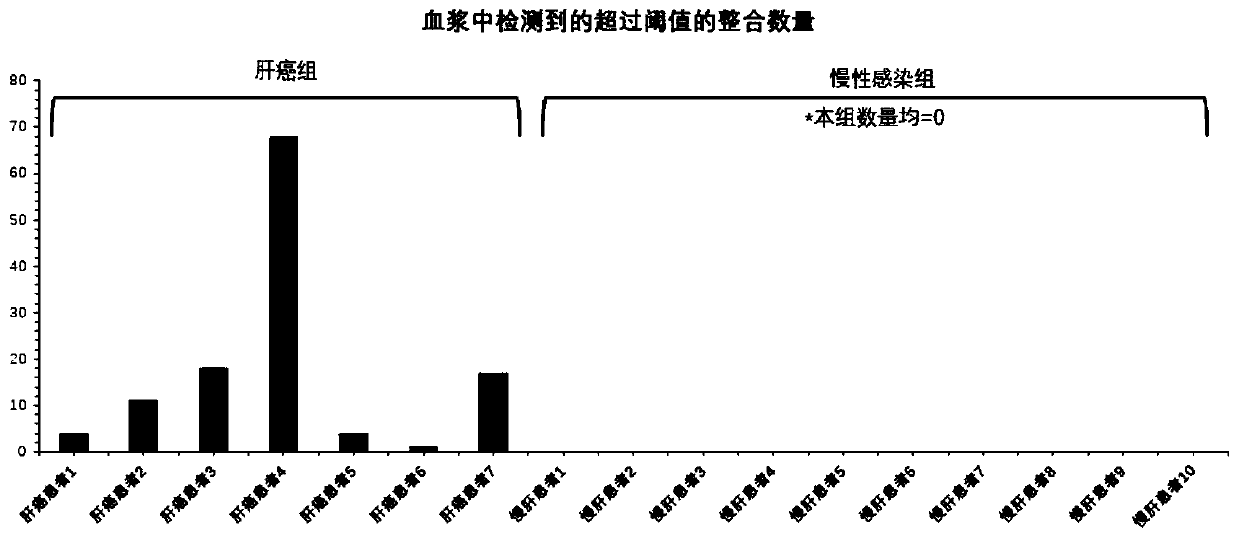
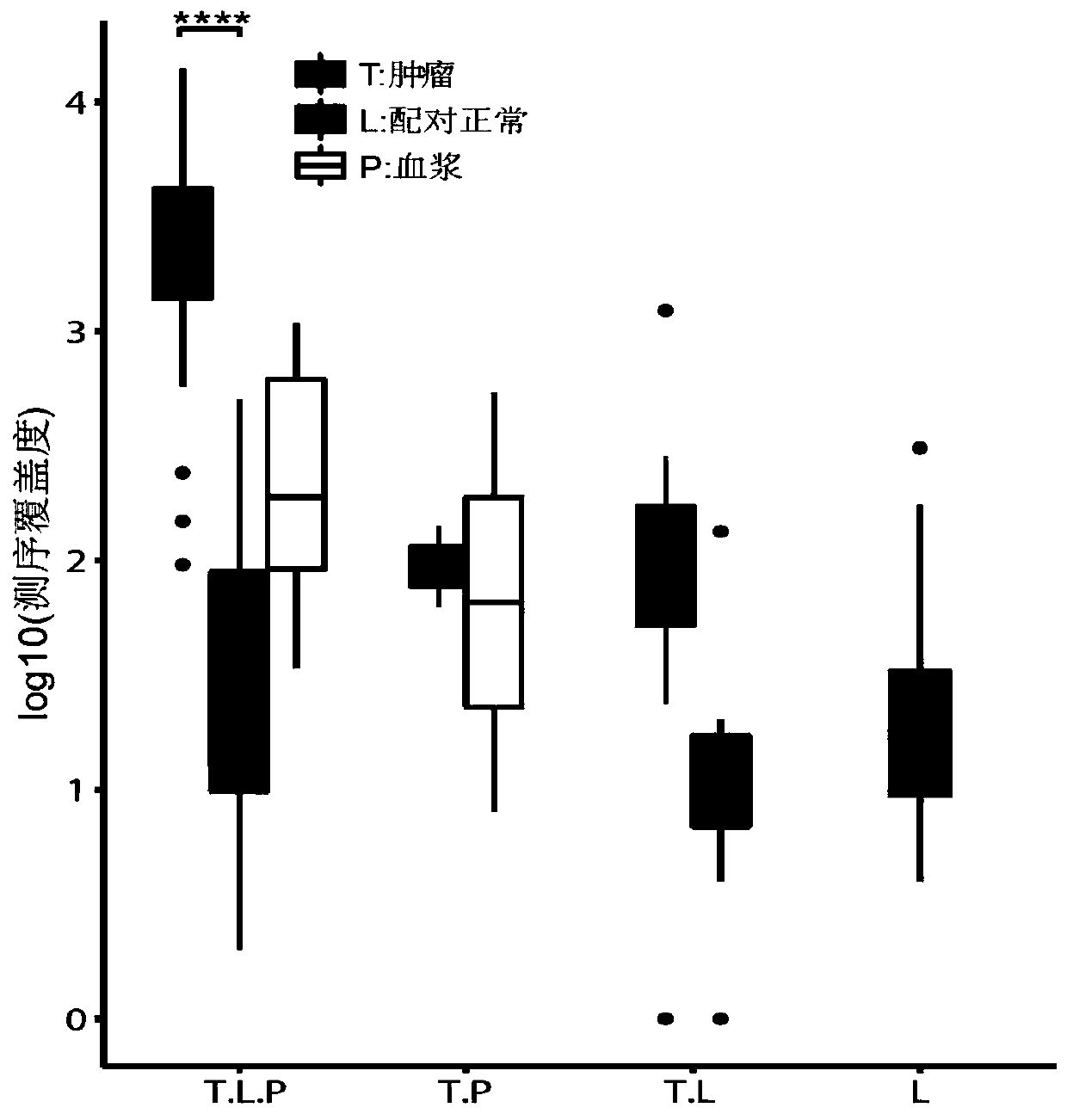
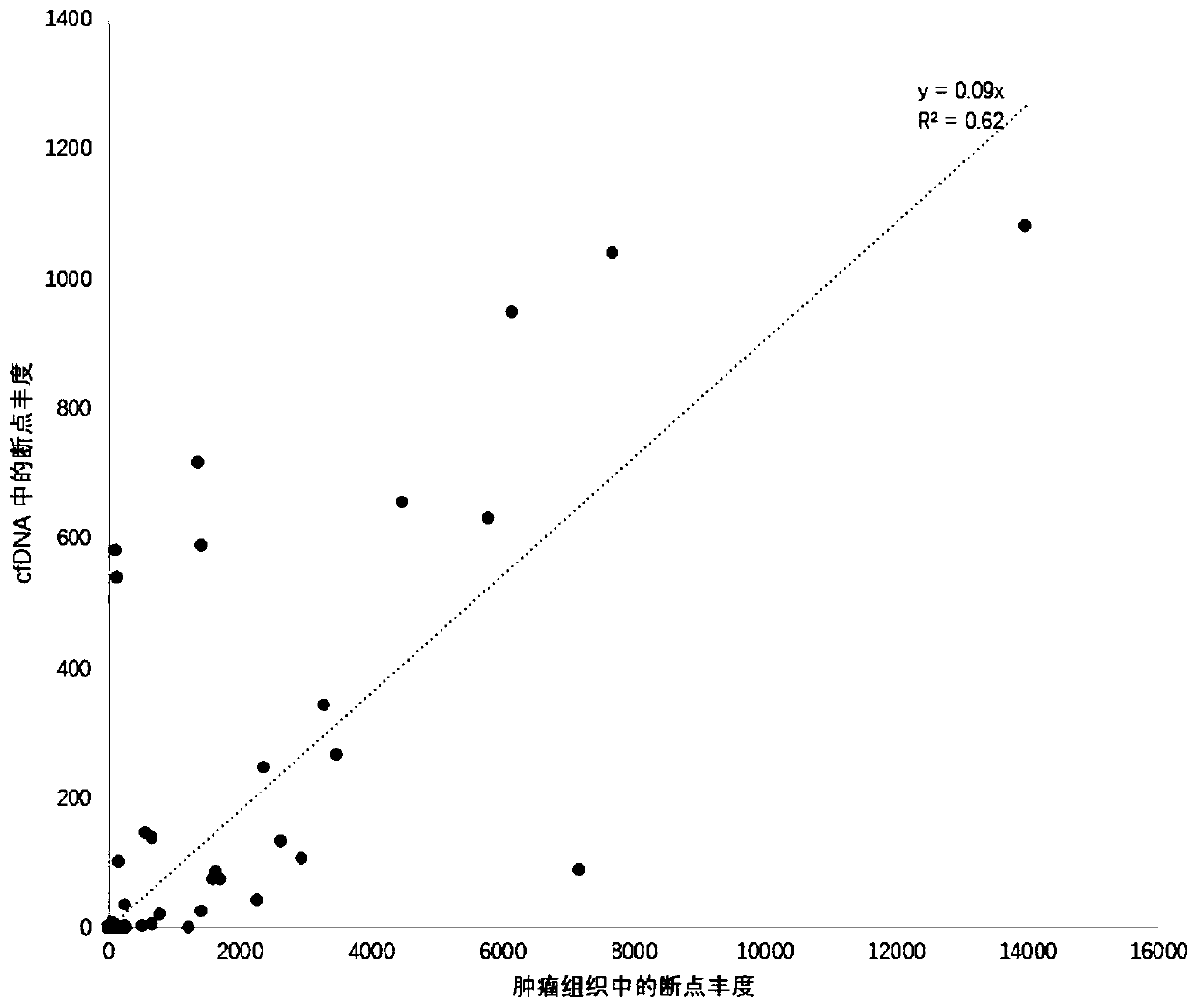
Peripheral blood detection method of hepatitis B virus integration in liver
A hepatitis B virus, peripheral blood technology, applied in biochemical equipment and methods, microbial determination/examination, etc., can solve problems such as limited information, no research to explore specificity and sensitivity, etc., to avoid complications, and to have good specificity. , the effect of avoiding heterogeneity bias
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1, sample collection and extraction of peripheral blood free DNA
[0077] Peripheral blood samples of 7 HCC patients after HBV infection were collected in Beijing You'an Hospital. For each liver cancer patient, in addition to collecting peripheral blood samples before surgery, 2 liver cancer tissue samples and 2 para-cancerous liver tissue samples were collected after surgery to control the ability to detect viral integration events in peripheral blood . The above-mentioned patients included 3 females and 4 males, with an average age of 51 years.
[0078]cfDNA from peripheral blood samples and DNA from liver cancer tissue samples and paracancerous liver tissue samples were extracted. DNA was extracted using the QIAGEN DNeasy Blood&Tissue Kits kit, and extracted according to the standardized procedure of the kit. The following steps are used for the extraction of cfDNA, with quantitative and quality assessments:
[0079] 1) Use free DNA blood collection tubes...
Embodiment 2
[0084] Example 2, End Repair, Adapter Ligation and Preamplification of Peripheral Blood Free DNA
[0085] The cfDNA obtained in Example 1 was processed according to the following steps:
[0086] 1) Quantify the extracted cfDNA to a volume of 50ul with ultrapure water, add 7ul of End Repair & A-Tailing Buffer and 3ul of End Repair & A-Tailing Enzyme Mix, mix to 60ul, Complete the end repair reaction on a thermal cycler, the reaction conditions are: 20°C for 30 minutes, 65°C for 30 minutes, store at 4°C, and keep the heating lid at 70°C.
[0087] 2) Add the above 60ul products to 5ul adapters (the adapters include primer regions for subsequent amplification and individual tag recognition sequences, which correspond to the adapters required by the subsequent sequencing platform), and perform ligation reaction on ice: add 55ul ligation reaction mixture , including 5ul nuclease-free water, 30ul ligation buffer, 10ul DNA ligase. Incubate for 15 minutes at 20°C (without heated lid)...
Embodiment 3
[0092] Embodiment 3, hybrid capture experiment
[0093] The integration event capture of peripheral blood cfDNA and liver cancer tissue and paracancerous tissue DNA is carried out as follows:
[0094] 1) Design of virus integration capture probe set: According to the sequences of hepatitis B virus A-H types obtained in the NCBI database, including 212 pieces of B type and 457 pieces of C type that are prevalent in China, design a degeneracy covering 3.2K of the whole virus genome Probes, each probe is about 100bp in length. The design method of the probe is well known in the art, and it can be completed by using a conventional design method, such as common software or a website. The probe set in this example is specifically designed through a commonly used primer design website https: / / design.igenetech.com / design obtained.
[0095] 2) Prepare the reaction solution: take the peripheral blood cfDNA dry powder sample obtained in Example 2, add nuclease-free water to quanti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com