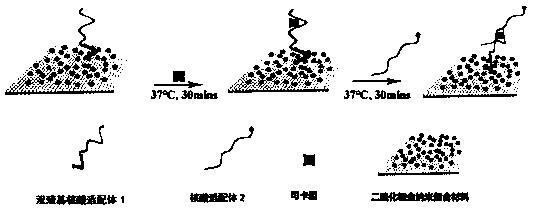
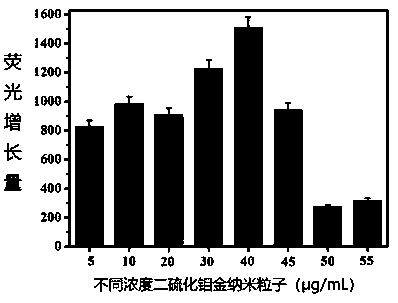
Method for detecting cocaine based on dimercapto aptamer
A technology of bis-mercapto nucleic acid and nucleic acid aptamer, which is applied in biological tests, measuring devices, material inspection products, etc., can solve the problems of cumbersome operation process, weak system stability, poor portability, etc., and achieves simple operation process and system stability. , easy to carry effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] (1) Synthetic specific DNA sequence:
[0046] DNA 1, DNA 2, and nucleic acid aptamer 2 were obtained using an exponential enrichment screening system and prepared by Shanghai Bioengineering Co., Ltd. The nucleotide sequences of DNA1, DNA2, and nucleic acid aptamer 2 are as follows:
[0047] The nucleotide sequence (SEQ.ID.NO.1) of DNA1 is: 5'-SH-CCATAGGGAGACAAGGATAAATCCTTCAATGAAGTGGGTCTCCC-3';
[0048] The nucleotide sequence (SEQ.ID.NO.2) of DNA 2 is: 3'-SH-GGTATCCCT-5';
[0049] The nucleotide sequence (SEQ.ID.NO.5) of the nucleic acid aptamer 2 is: 5'-CCATAGGGAGACAAGATAAATCCTTCAATGAAGTGGGTCTCCC-FAM-3';
[0050] Among them, DNA1 has no fluorescence, and a sulfhydryl group is modified at the 5' end; DNA 2 has no fluorescence, and a sulfhydryl group is modified at the 3' end, which is partially complementary to DNA 1; FAM in nucleic acid aptamer 2 is carboxyfluorescein, which is a fluorescent modification group. The fluorescent group is modified at the 3' end without ...
Embodiment 2
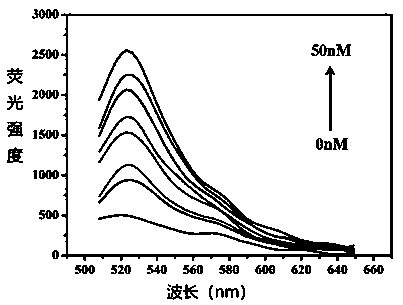
[0076] In this example, the concentration of cocaine and its analogs adenosine, uridine, and uric acid in the specificity verification experiment of step (7) in Example 1 was changed to 200nM, and the concentration of cocaine in human serum was added to step (8). Change to 0.05nM, 0.5nM, 50nM; other steps are the same as in Example 1.
[0077] The experimental result of this embodiment is:
[0078] (1) Established fluorescence growth rate F / F 0 The linear regression equation between -1 and cocaine concentration x is still y=12.28497x+0.58782 with embodiment 1, R 2 =0.96752, where y represents the fluorescence growth rate F / F 0 -1. According to the 3S / N principle, the calculated detection limit of cocaine is about 0.54pM.
[0079] (2) The fluorescence signal of cocaine is the highest in the specificity verification experiment, which shows that only cocaine can cause the fluorescence of the detection system to be significantly enhanced, and also shows that the detection meth...
Embodiment 3
[0083] In this example, the water bath condition in step (2) of Example 1 was changed to 4°C for half an hour, and other methods and steps were the same as in Example 1.
[0084] Experimental results:
[0085] (1) Established fluorescence growth rate F / F 0 The linear regression equation between -1 and cocaine concentration x is y=11.78529x+0.89573, R 2 =0.97846, where y represents the fluorescence growth rate F / F 0 -1, the calculated detection limit for cocaine is about 0.54pM.
[0086] (2) The fluorescence signal of cocaine is the highest in the specificity verification experiment, which shows that only cocaine can cause the fluorescence of the detection system to be significantly enhanced, and also shows that the detection method designed in the present invention has obvious selectivity.
[0087] (3) The cocaine concentration actually detected in human serum is close to the concentration of 0.01nM, 0.1nM, and 10nM added before the detection. For reliable data), the recov...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap