Barley hvals1 gene and its use
A barley and gene technology, applied in the field of genetic engineering, can solve the problems of few aluminum-resistant genes, sensitivity to aluminum hordeate stress, and few reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Cloning and Analysis of CDS Region and Promoter Sequence of HvALS1 Gene
[0034] 1. Cloning of CDS region sequence of HvALS1 gene
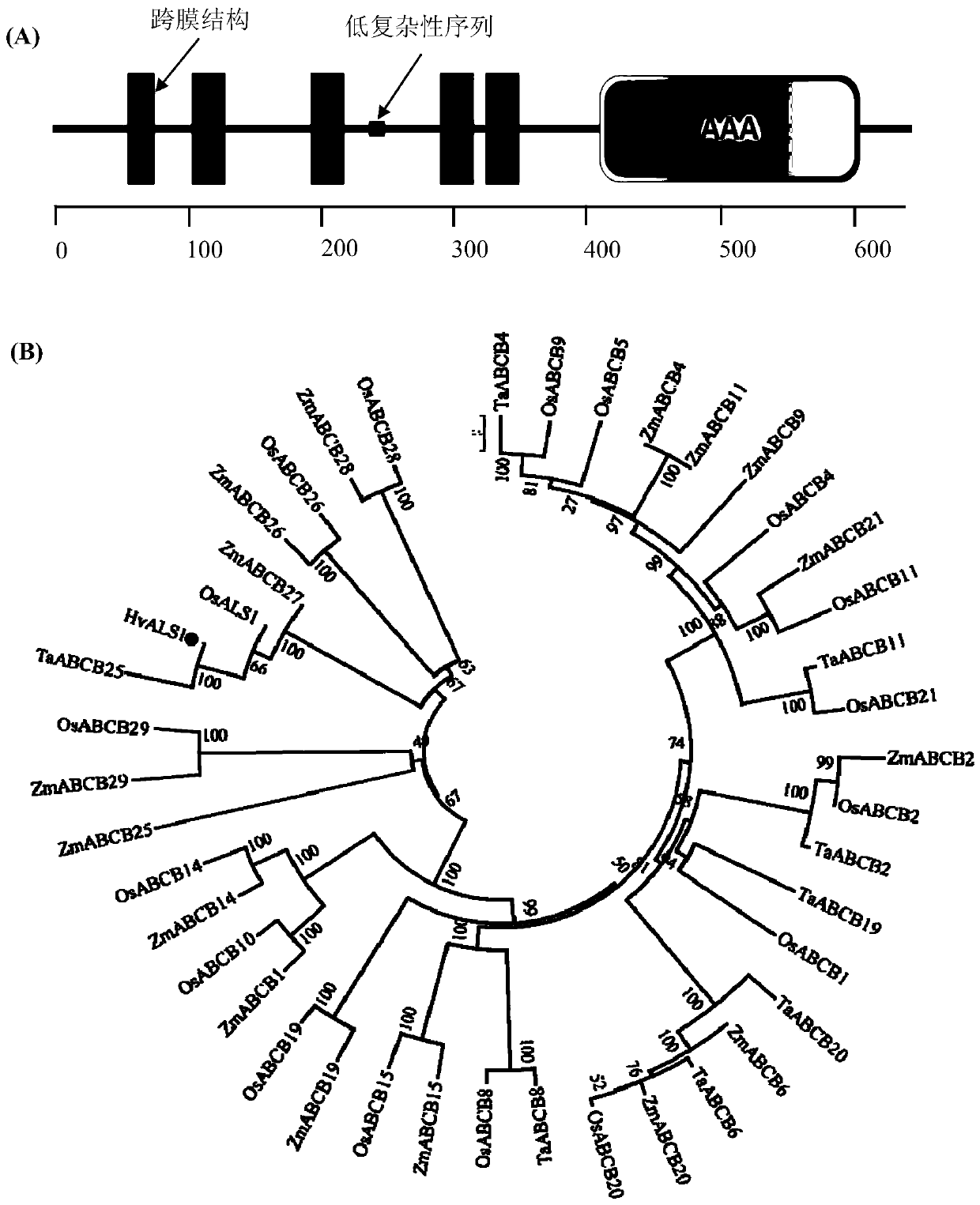
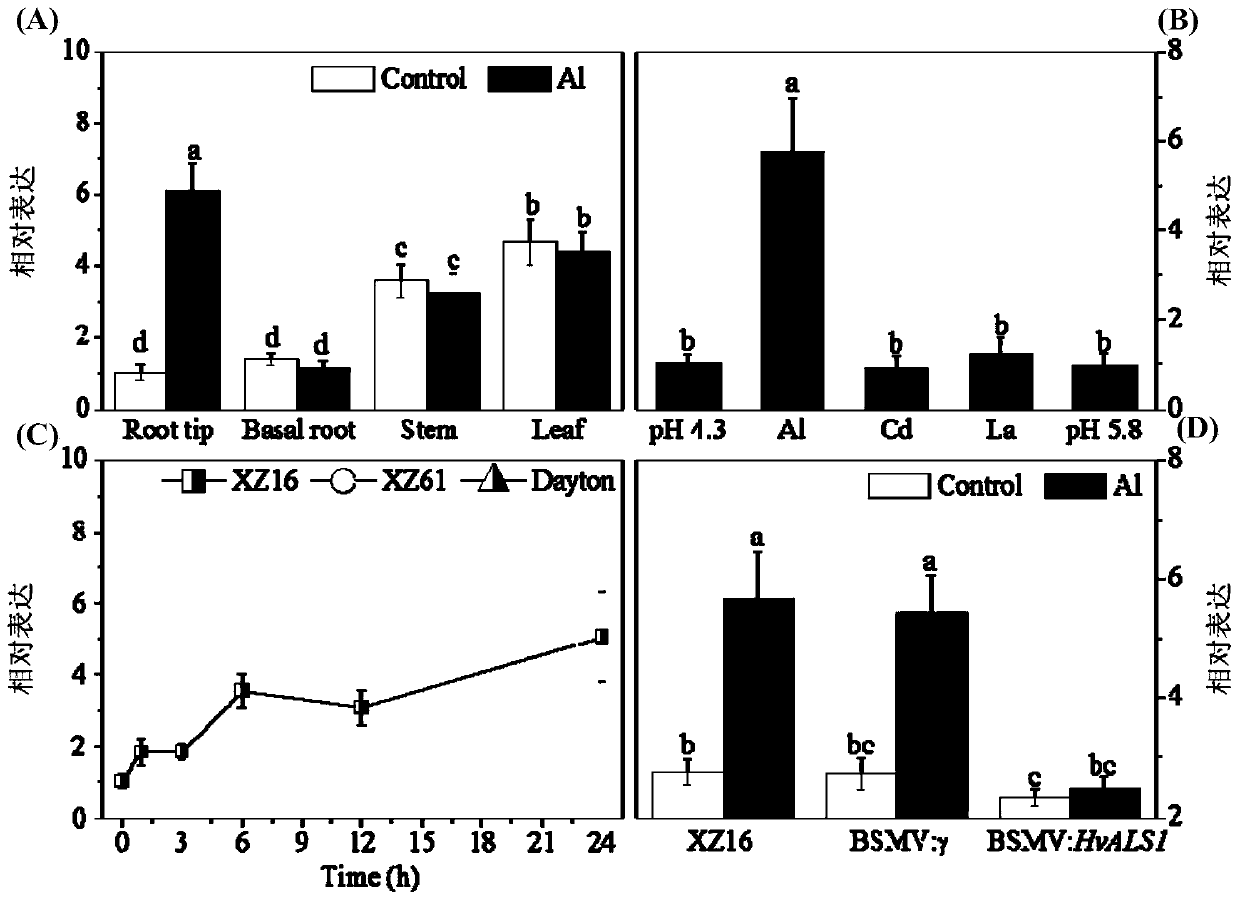
[0035] Based on the previous research of the research group, a differentially expressed gene in the Al-tolerant genotype (XZ16) and Al-sensitive genotype (XZ61) under Al stress conditions was selected from the root gene chip structure, and a highly expressed ABC in the Al-tolerant genotype was selected. Family gene, the full-length CDS region sequence of this gene was cloned from XZ16 (as shown in SEQ ID NO. 1), and it was named as HvALS1.
[0036] Total RNA from the roots of annual wild barley XZ16 on the Qinghai-Tibet Plateau was extracted using a total RNA extraction kit (Takara, Japan), and genomic DNA contamination in the total RNA was removed with DNaseI (Takara, Japan). PrimeScripffM II 1stStrand cDNA Synthesis Kit reverse transcription kit ( Takara, Japan) reverse transcribed the extracted total RNA into single-stranded cDNA. Desi...
Embodiment 2
[0056] Verification of HvALS1 gene function by BSMV-VIGS method
[0057] 1. Construction of BSMV:HvALS1 vector
[0058] Total RNA from XZ16 roots was extracted using a total RNA extraction kit (Takara, Japan), and genomic DNA contamination in the total RNA was removed with DNaseI (Takara, Japan), using PrimeScript TM II 1st Strand cDNA Synthesis Kit Reverse Transcription Kit (Takara, Japan) reverse-transcribes total RNA into cDNA. Primers were designed according to the results of Blast, and a 262bp HvALS1 gene fragment was obtained by amplification.
[0059] KOD enzyme PCR reaction system:
[0060]
[0061] The amplification program was: 94°C for 5 min, (98°C for 10s, 58°C for 30s, 68°C for 30s) for 35 cycles, and 68°C for 10 min.
[0062] The primer sequences are (underlined are the restriction sites):
[0063] HvALS1-γ-F: 5'-GTAC GCTAGC CAAAACATGACGCCGGGAAG-3' (SEQ ID No: 10);
[0064] HvALS1-γ-R: 5'-GTAC GCTAGC CGAGCAACGACTCTCTCACT-3' (SEQ ID No: 11).
[0065] ...
Embodiment 3
[0083] HvALS1 gene overexpression
[0084] 1. Construction of overexpression vector
[0085] The method for constructing the overexpression vector in this experiment is Gateway. Total RNA of barley variety Golden Promise was extracted, reverse transcribed into cDNA, and primers for overexpression vector of HvALS1 gene were designed, and cDNA was used as template to amplify the target gene. After sequence alignment, the CDS sequence of the Golden Promise HvALS1 gene was identical to that of XZ16.
[0086] Wherein the overexpression primer of HvALS1 gene amplifies the target fragment primer sequence of 1935bp as follows:
[0087] AttB-F-Overexpression-HvALS1:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGTGAGGGAGCTGCGC (SEQ ID No: 13);
[0088] AttB-R-Overexpression-HvALS1:GGGGACCACTTTGTACAAGAAAGCTGGGTCTATTGGCCATTACTGCTGG (SEQ ID No: 14).
[0089] The position of the bands was detected by 1% agarose gel electrophoresis on the products amplified by PCR, the target gene was recovered and ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com