Application of a bayberry CAPS marker in detection of allo-related MrMYB1 Alleles in Chinese Bayberry
An allele and marker technology, applied in the field of CAPS markers, can solve the problems of time-consuming, laborious and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: CAPS mark development ideas
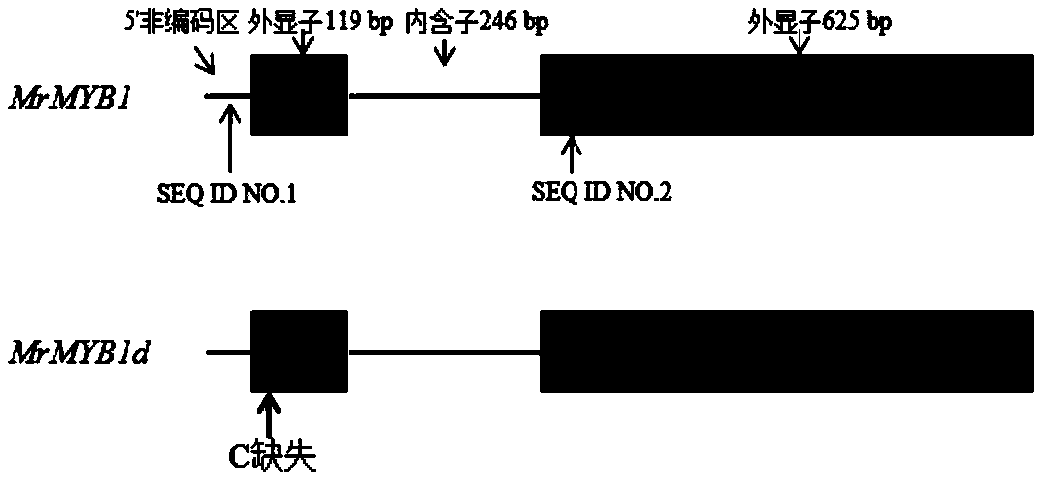
[0029] The research proved that MrMYB1 gene sequence contains 2 exons, 119bp and 625bp respectively, and 1 intron, 246bp. The allele MrMYB1d is missing a C at the 30th position after the start codon (ATG) (attached figure 1 ), resulting in a frameshift mutation that cannot synthesize normal MrMYB1 protein. It was found that this deletion mutation caused the loss of a HhaI recognition site (5'-GCGC-3') of the sequence, thus enabling the development of CAPS markers.
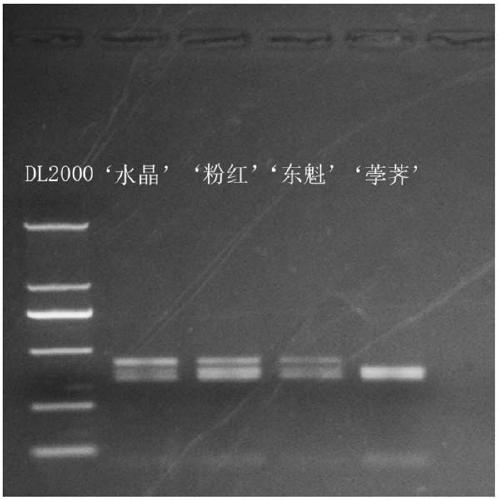
[0030] In order to make the PCR product of allele MrMYB1 produce well-separated bands after HhaI digestion and be applicable to both genomic DNA and cDNA, the selected upstream and downstream primers should have different distances from the restriction site, and the distance between the upstream and downstream primers should be different. No additional HhaI cleavage site should be present. Considering the above two requirements, the upstream primer was finally des...
Embodiment 2
[0031] Embodiment 2: CTAB method extracts Waxberry genome DNA
[0032] (1) Prepare CTAB genomic DNA extract, the formula is: 2% CTAB (hexadecyl triethyl ammonium bromide, Hexadecyl trimethyl ammonium bromide), 0.1M Tris, 20mM EDTA, 1.4M NaCl, pH 8.0.
[0033] (2) Collect the young leaves of red bayberry.
[0034] (3) Put it into liquid nitrogen and grind it into powder, weigh about 0.7g and transfer it into a 10ml centrifuge tube filled with 4ml CTAB solution and 80μL β-mercaptoethanol (preheated at 65°C), bathe in 65°C water for 1h; add 4ml Chloroform / isoamyl alcohol (24:1, v / v), mix well, centrifuge at 12000rpm for 10min, 15°C, take supernatant and add 4ml chloroform / isoamyl alcohol (24:1, v / v) again, mix well , centrifuged at 12000rpm for 10min. Aspirate the supernatant, add 2ml 5M NaCl, 4ml isopropanol (pre-cooled at -20°C), mix well, and place at -20°C for 2h. Centrifuge at 12000rpm for 15min, discard the supernatant, rinse the precipitate twice with 75% ethanol, and d...
Embodiment 3
[0035] Embodiment 3: RNA extraction and cDNA synthesis of red bayberry fruit
[0036](1) Prepare CTAB RNA extract, formula is: 2% CTAB, 2% PVP-K30 (vinylpyrrolidone iodine, polyvinylpyrrolidone), 100mL 1M Tris, 50mL 0.5M EDTA, 2M NaCl, 0.5% spermidine, pH 8.0.
[0037] (2) Gather ripe red bayberry fruit.
[0038] (3) Extract RNA. Put the bayberry fruit into liquid nitrogen and grind it into powder, weigh about 1.0g and transfer it into a 10mL centrifuge tube filled with 4mL CTAB solution and 80μL β-mercaptoethanol (preheated at 65°C), bathe in 65°C water for 5min; add 4ml chloroform / Isoamyl alcohol (24:1, v / v), mix well, centrifuge at 10000rpm for 10min, take the supernatant and add 4ml of chloroform / isoamylalcohol (24:1, v / v), mix well, and centrifuge at 12000rpm for 10min. Aspirate the supernatant, add 2M LiCl, mix well, and place at 4°C overnight. Centrifuge at 10,000 rpm for 20 min at 4°C, discard the supernatant, dissolve the precipitate with SSTE at 65°C, add 400 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com