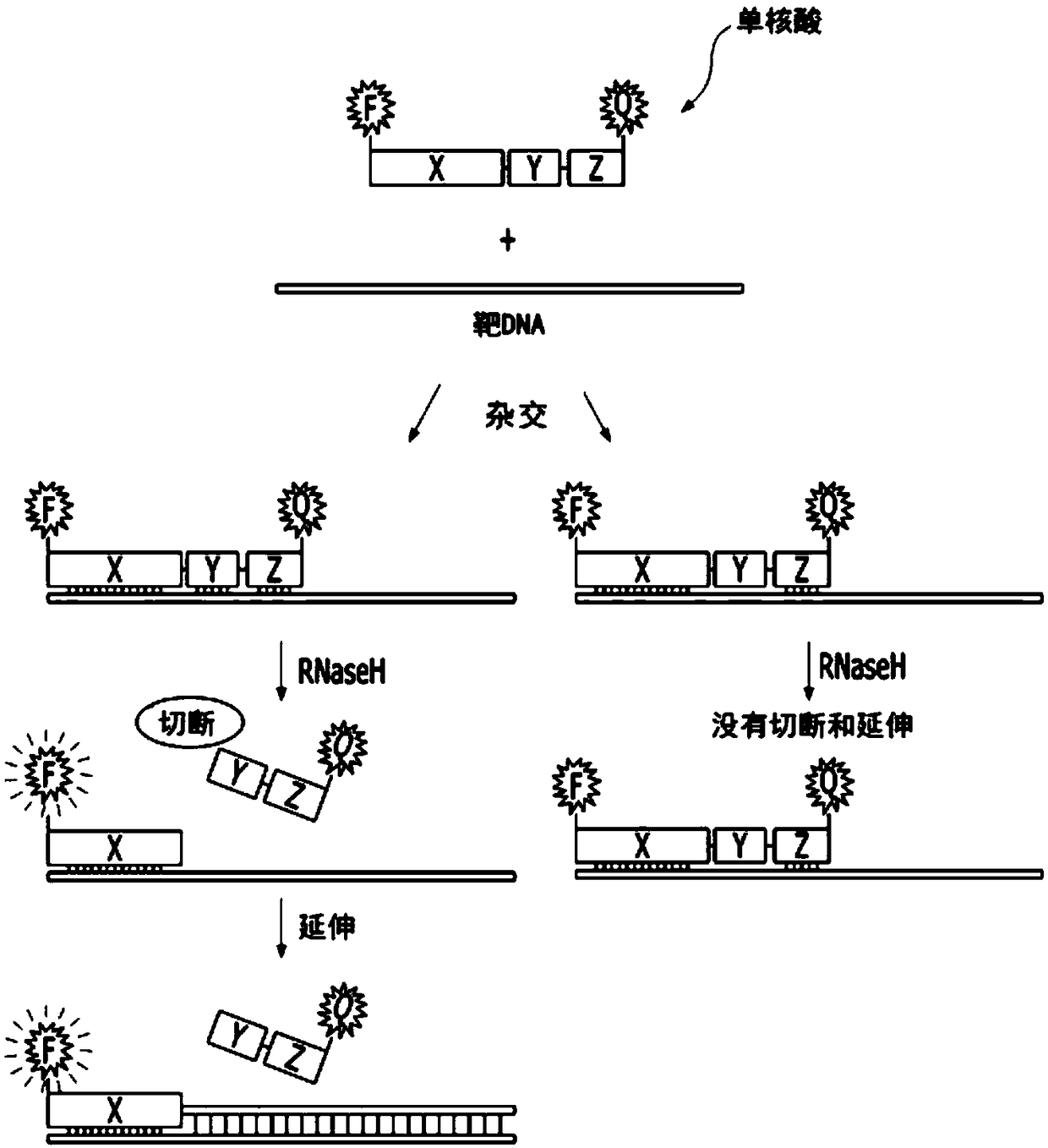
Single-stranded nucleic acid for detecting nucleic acid or protein in real time and detection method using same
A real-time detection, single-nucleic acid technology, applied in biochemical equipment and methods, measurement devices, and microbial determination/inspection, etc., can solve the problems of difficult to analyze SNP, difficult to apply endpoint genotyping, reduced accuracy, etc., and achieve savings. Analyze the effects of time and expense
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
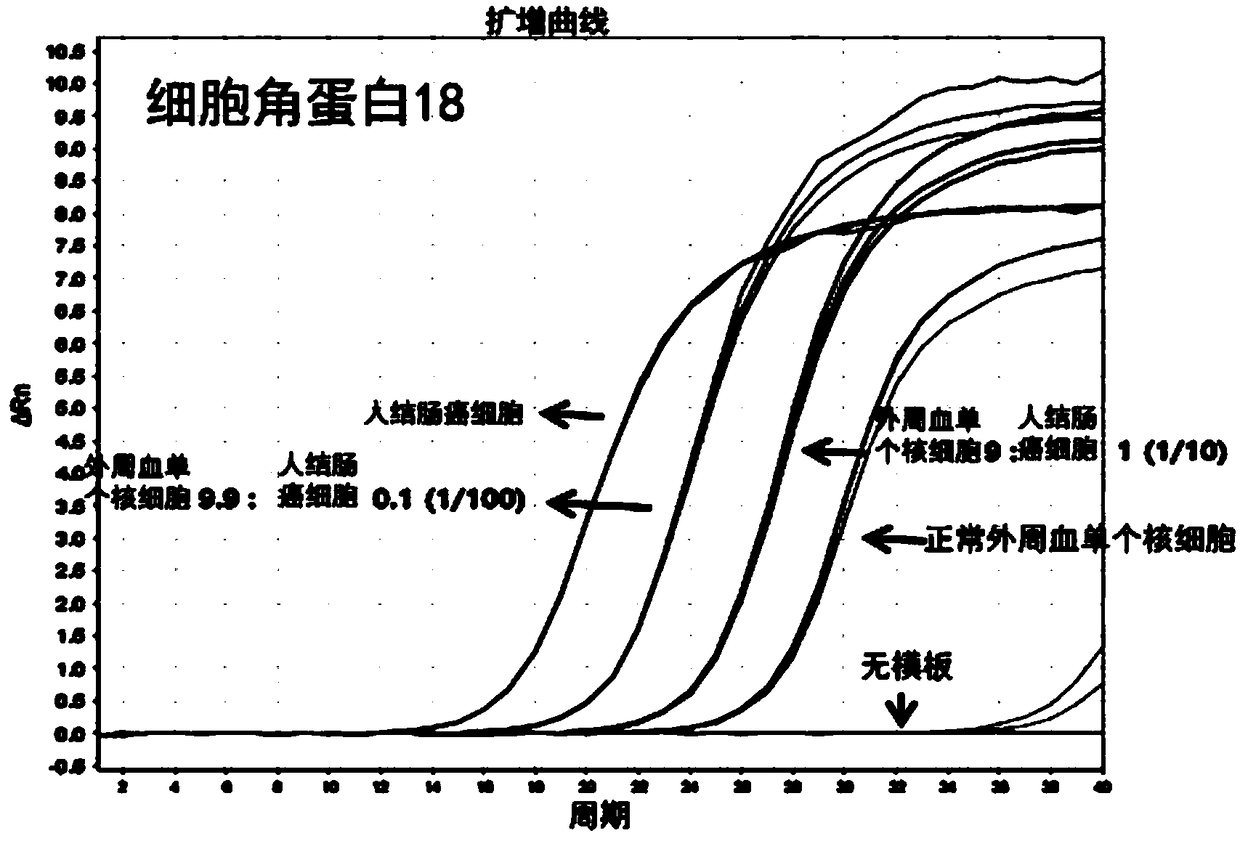
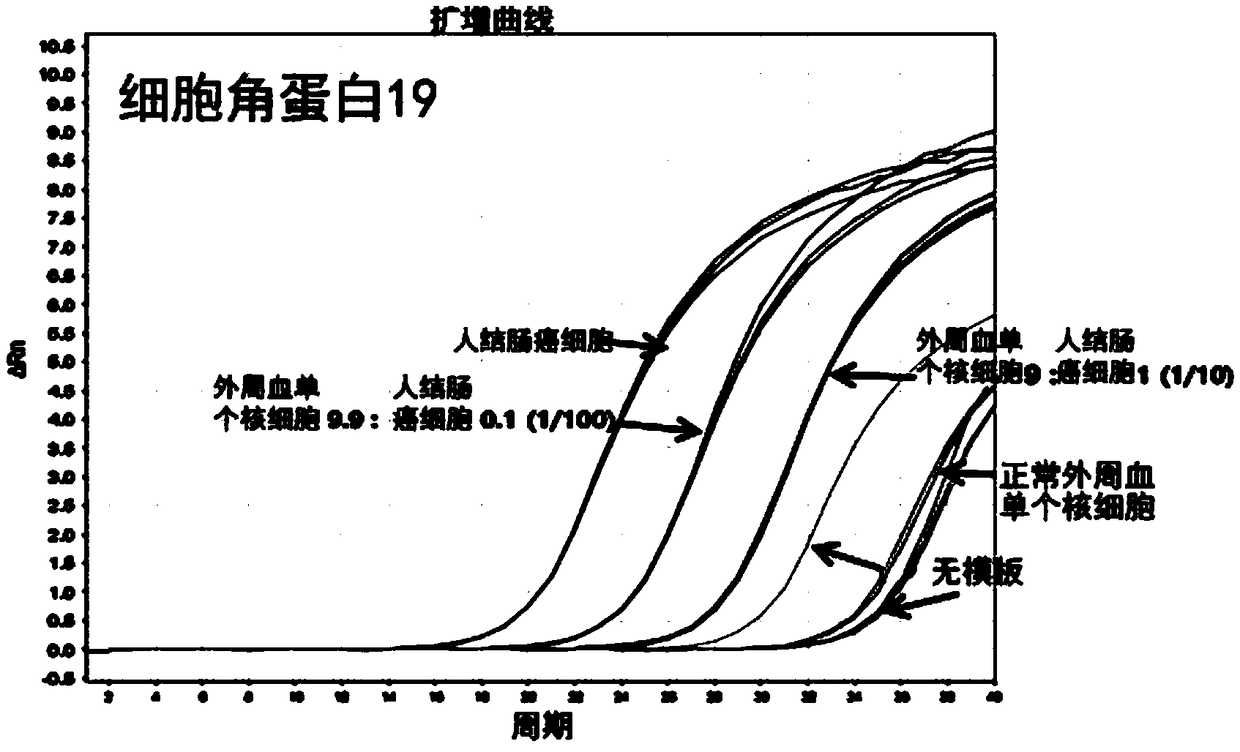
[0169] Embodiment 1: Utilize the real-time nucleic acid analysis of single nucleic acid of the present invention
[0170]In order to measure cytokeratin-18 (CK-18, cytokeratin-18), cytokeratin-19 (CK-19, cytokeratin-19) and 3-phosphate glyceraldehyde dehydrogenase (GAPDH, Glyceraldehyde 3-phosphatedehydrogenase) Whether the gene is expressed, as described in Table 1 below, the single nucleic acid (promer) and primer (primer) of the present invention were entrusted to Integrated DNA Technologies (IDT, Integrated DNA Technologies, USA) for manufacture (refer to Table 1). Among them, for a single nucleic acid (promer), fluorescein succinimidyl ester (FAM, fluoresceinsuccinimidyl ester) is attached to the 5' end, and 3IABkFG is attached to the 3' end. In order to distinguish ribonucleotide (RNA) from deoxyribose nucleus Nucleotide (DNA), preceded by the sequence, is indicated by the subscript "r". As a comparison group, the primer to be used in the SYBR method was entrusted to ID...
Embodiment 2
[0177] Example 2: Mutation assay using single nucleic acid of the present invention
[0178] Using the single nucleic acid (promer) of the present invention, it is determined whether G12D and G12V of the KRAS gene (gene) are mutated. Specifically, entrusted to IDT, the single nucleic acid (promer) and reverse primer (reverse primer) of the present invention were manufactured according to the following Table 3. For the single nucleic acid (promer), fluorescein succinyl was attached to the 5' end. Fluorescein succinimidyl ester (FAM, fluorescein succinimidyl ester), with 3IABkFG attached at the 3' end, in order to distinguish ribonucleotide (RNA) and deoxyribonucleotide (DNA), in front of the sequence, it is indicated by subscript "r".
[0179] table 3
[0180]
[0181]
[0182] In addition, total DNA was obtained from SW620 (BioSample: SAMN05292430) and LS174T cell line (BioSample: SAMN03701686) for the KRAS gene to be measured. (Please inform NCBI of the source.) The w...
Embodiment 3
[0188] Example 3: Optimal Size Determination of Single Nucleic Acids of the Invention
[0189] The OPN1MW gene (Themedium-wave-sensitive opsin-1 gene, Themedium-wave-sensitive opsin-1gene) located on the long arm of the X chromosome (Xq28) is a gene related to color weakness and color blindness, and mutations occur at 4 nucleotide positions , thus being found to be red-green weak. The mutation positions are C282A, T529C, T607C, and G989A.
[0190] Accordingly, in order to confirm whether the mutation based on the length of the single nucleic acid (promer) of the present invention was detected, the present inventors produced a DNA sequence inserted into the 241st to 1030th nucleotide (see the DNA sequence of SEQ ID NO: 13) of the OPN1MW T529C mutant gene. After plasma, after dilution at concentrations of 10fM, 1fM, 100aM, and 10aM, each 1 μl was removed, and as shown in Table 4 below, five kinds of single nucleic acids and reverse primers manufactured by IDT were used for dete...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com