Deinococcus radiodurans DNA transmethylase
A technology of Deinococcus radiodurans and methyltransferase, applied in transferase, enzyme, enzyme and other directions, can solve the problems of M.DraR1 modification mode and substrate recognition sequence that have not been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1: Deinococcus radiodurans DNA methyltransferase M.DraR1 substrate recognition specificity
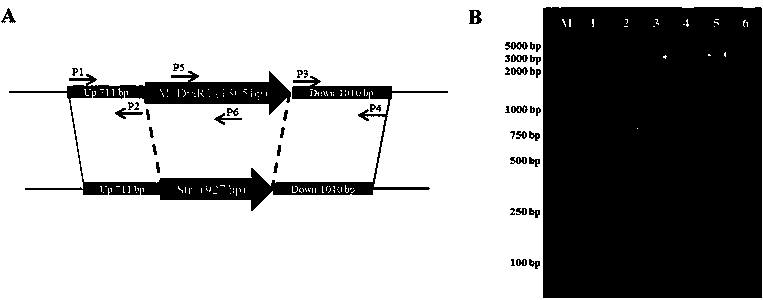
[0020] (1) DNA methyltransferase M.DraR1 Construction of gene deletion strains: using the classic "three-segment linking method" to M.DraR1 The upstream fragment (about 710 bp) and downstream fragment (about 1000 bp) of the gene sequence and the resistance gene fragment (streptomycin resistance gene Str, about 927 bp) are linked into a whole fragment in vitro by T4 ligase (Takara company) (See figure 1 A), and then transform this triple product into competent cells of Deinococcus radiodurans, screen the mutant strains on the Str-resistant TGY medium plate, and obtain the result by PCR and sequencing verification M.DraR1 Gene deletion strain—Δ M.DraR1 (See figure 1 B).
[0021] (2) Deinococcus radiodurans wild-type DraR1 and M.DraR1 Extraction of gene knockout strain genome: pick wild-type DraR1 and knockout strain Δ M.DraR1 Inoculate a single colony into 5 mL of...
Embodiment 2
[0027] Example 2: In vivo enzyme activity analysis of DNA methyltransferase M.DraR1
[0028] (1) Construction of the in vivo expression vector pRRS-M.DraR1 vector: using the genomic DNA of Deinococcus radiodurans as a template, using pfu High-fidelity polymerase (full gene, AP 221) was used to amplify the M.DraR1 gene in vitro by PCR. The upstream primer pRRS-M.DraR1-F contained the Sbf I restriction site, the TTAAGG box and the TTAATCAT sequence. The sequence is shown in SEQ ID NO. 2; the downstream primer pRRS-M.DraR1-R contains a BamH I restriction site and a CCGCGG substrate conservative sequence, and its sequence is shown in SEQ ID NO. 3. 1.0% agarose electrophoresis detection, purification of PCR products (Life company DNA purification kit, Cat No. 116401), purified PCR fragments and pRRS plasmid (gifted by Professor Roberts of NEB company) and Sbf I and BamH I (NEB company) at the same time Carry out enzyme digestion, purify the digested fragments, connect them with T...
Embodiment 3
[0032] Example 3: Analysis of DNA methyltransferase M.DraR1 in vitro enzyme activity
[0033] (1) Linearization treatment of pRRS-M.DraR1 plasmid: Cut the above obtained plasmid into linear fragments with Hind III, purify and recover for later use.
[0034] (2) DNA methyltransferase M.DraR1 in vitro reaction system: Add 1-2 μg of the above-mentioned treated plasmid into a solution containing 50-200 mM KCl, 10-50 mM Tris-HCl (pH7.5-8.0), 0.1 mM Add 1 μM purified M.DraR1 protein to the reaction buffer of EDTA, 3-7 mM β-Me and 20-100 μM SAM, and react under suitable conditions.
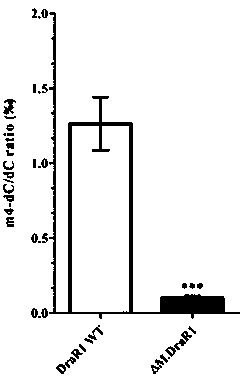
[0035] (3) Optimum temperature range for the enzyme activity of DNA methyltransferase M.DraR1: place the above reaction systems at 4-60 °C for 0.5-1 h, and purify the plasmid DNA or phage DNA after the reaction. The results further indicated that M.DraR1 methyltransferase can methylate and modify the "CCGCGG" conservative motif, and its optimum temperature range is 25-37 ℃. At 4 °C, it also has weak me...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com