Aptazyme sequence
A nucleic acid aptamer and ribozyme technology, applied in biochemical equipment and methods, instruments, analytical materials, etc., to achieve the effects of high sensitivity, improved sensitivity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
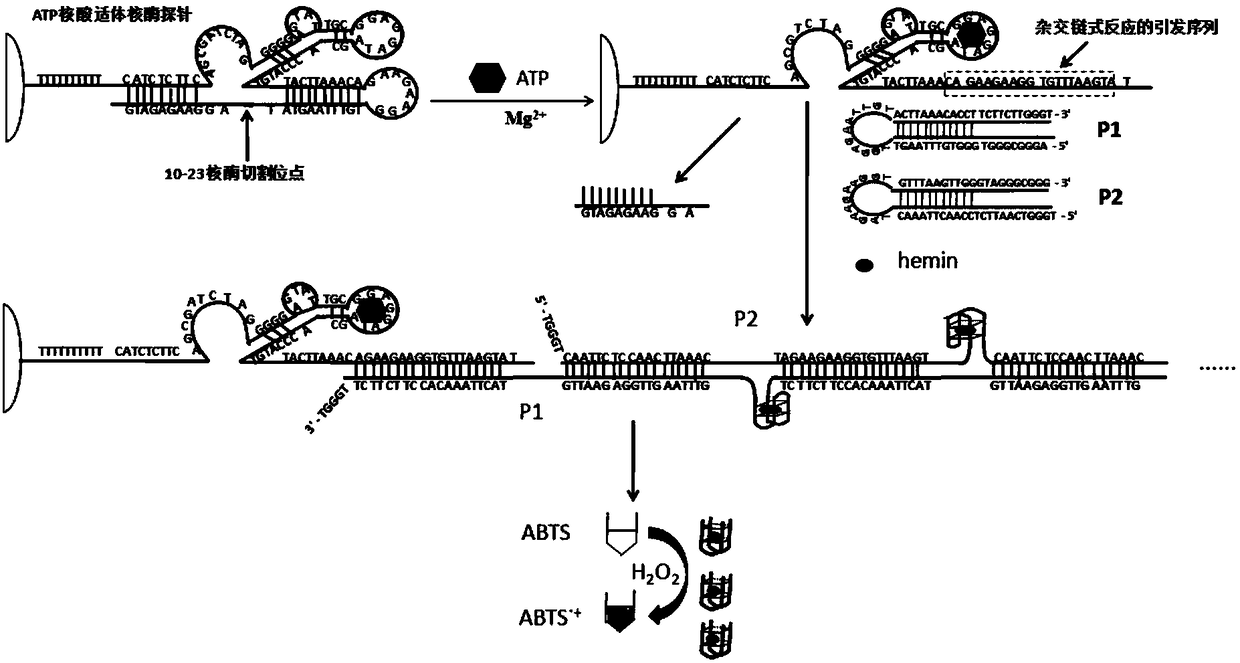
[0050] Embodiment 1 ATP nucleic acid aptamer ribozyme probe design
[0051] (1) The structure of the ATP nucleic acid aptamer ribozyme probe is as follows:
[0052]
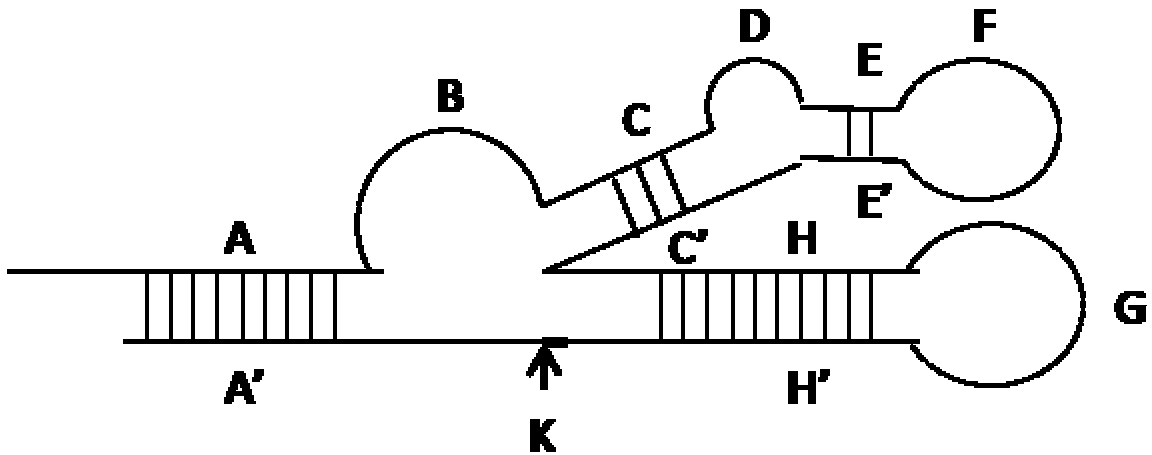
[0053] The first line "CATCTCTTC" and the second line "GAAGAGATG" (attached figure 1 Parts A and A' in the middle) complementarily hybridize to form the stem in the molecule; the second line "TACTTAAACA" and "TGTTTAAGTA" part (attached figure 1 The H and H' parts in the middle) complementarily hybridize to form the stem-loop G in the molecule; the first line "AGCGATCTA" (SEQ ID NO.1) is the 10-23 ribozyme sequence (attached figure 1 Middle part B), the first line "GGGGGAGTATTGCGGAGGA" (SEQ ID NO.2) part is the ATP nucleic acid aptamer sequence (attached figure 1 F in part), the second line "AGAAGAAGGTGTTTAAGTA" (SEQ ID NO.3) part (attached figure 1 Middle G and H' parts) are the priming sequences of the hybridization chain reaction. When there is no ATP, this sequence is closed in the stem of the molec...
Embodiment 2
[0060] Embodiment 2 is to the detection of different concentration ATP
[0061] (1) Formation of ATP aptamer ribozyme secondary structure
[0062] The biotin-modified aptamer ribozyme uses 25mM HEPES buffer (25mM HEPES, 100mM NaCl20mM MgCl 2 ,pH=7.4) configuration. In order to reduce the generation of non-hairpin dimers and increase the formation ratio of hairpin structures, stepwise slow annealing was adopted. Take 100 μL of 1 μM biotinylated aptamer ribozyme in an EP tube, and gradually anneal as follows: 95°C for 5 minutes, 60°C for 5 minutes, 40°C for 5 minutes, 30°C for 10 minutes, 20°C for 15 minutes, and store at 4°C.
[0063] (2) Formation of secondary structures of hairpin probes P1 and P2
[0064] Hairpin probes P1 and P2 use 25mM HEPES buffer (25mM HEPES, 100mM NaCl 20mM MgCl 2 ,pH=7.4) configuration. Take 100 μL of 0.5 μM hairpin probes P1 and P2 in EP tubes, and gradually anneal slowly in order to reduce the generation of non-hairpin dimers and increase the for...
Embodiment 3
[0070] The specificity of embodiment 3 ATP detection
[0071] In order to test the specificity of the sensing system for ATP detection, three other interfering molecules (UTP, GTP, CTP) were selected and their interference degree to ATP detection was tested respectively. The concentration of the four small molecule compounds is 1nmol / L. From the results, the detection signal of ATP is much greater than that of other interfering molecules. See the attached Figure 5 , which shows that the technical system has high specificity to ATP and can meet industrial clinical application.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com