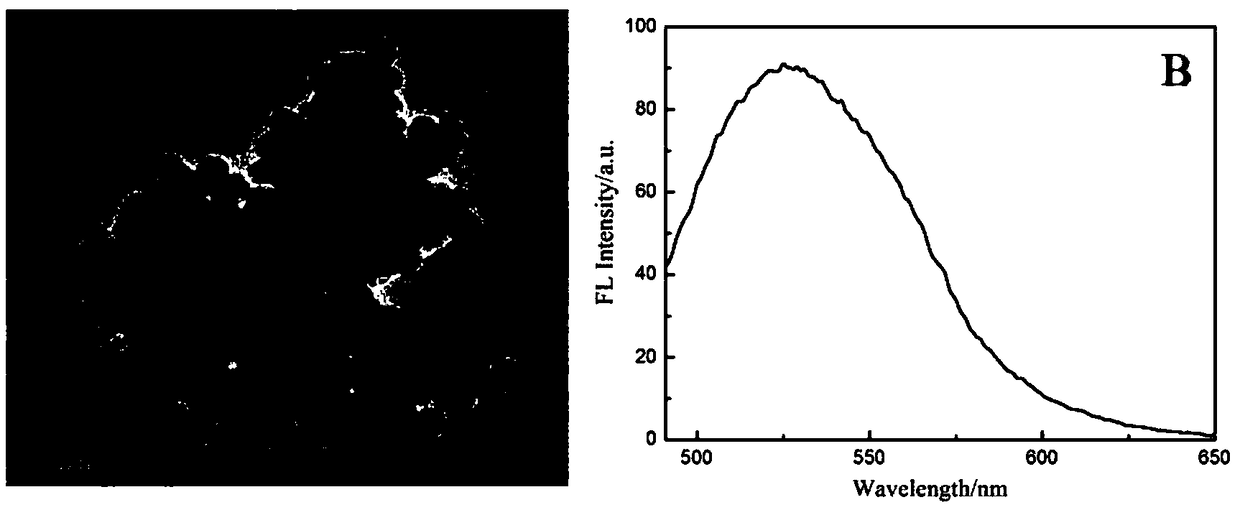
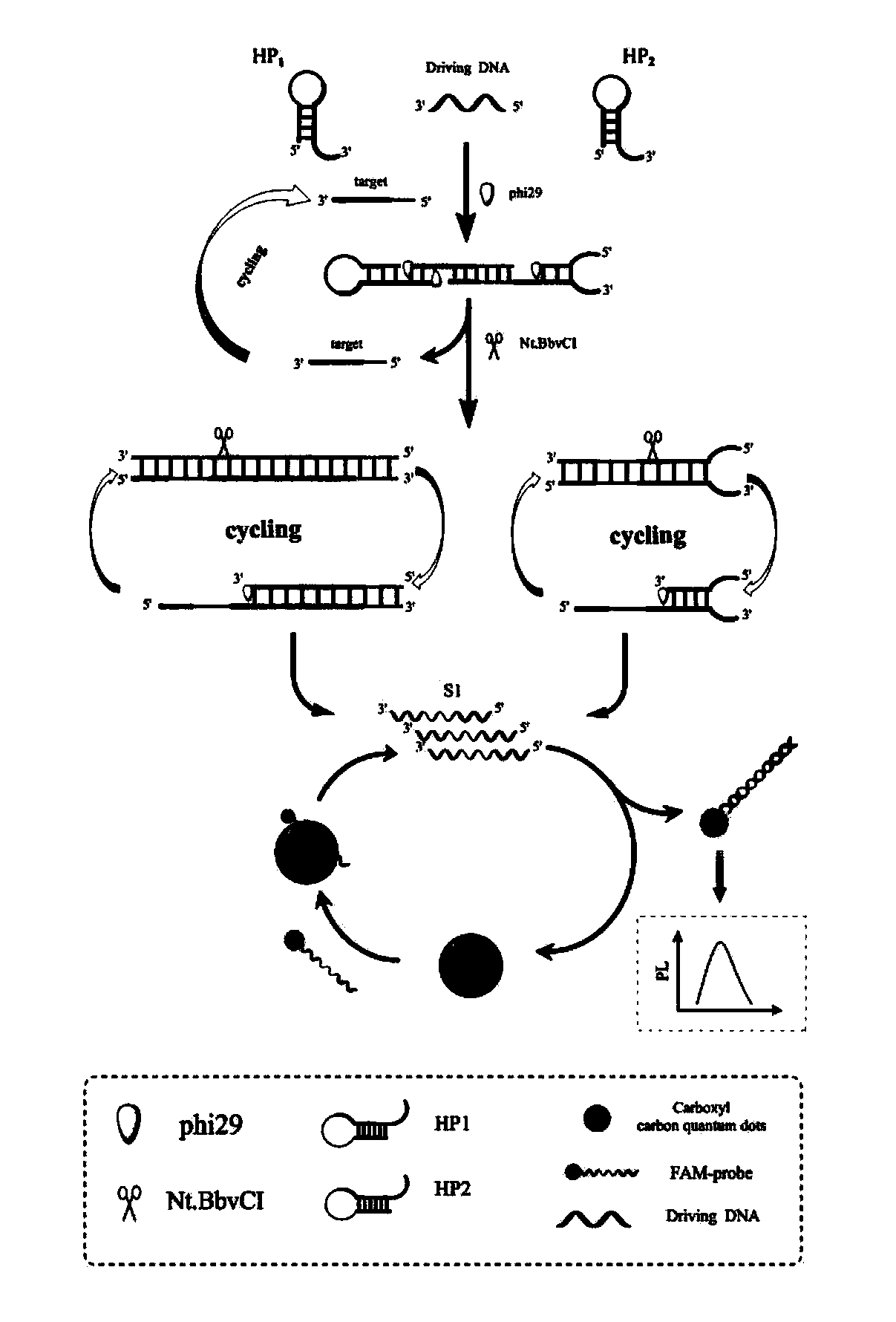
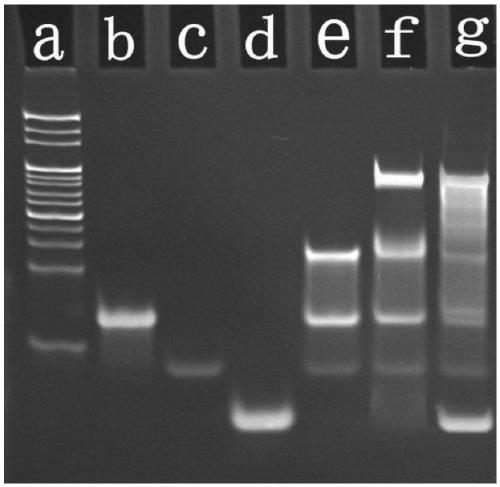
Fluorescent biosensor based on cyclic amplification technology and carboxyl carbon quantum dots as well as preparation method and application of fluorescent biosensor
A technology of biosensors and carbon quantum dots, applied in the field of biosensors, can solve the problems of complex and strict preparation of nanomaterials, difficulty in ensuring the biocompatibility of nanomaterials, etc., and achieve excellent accuracy, great application potential, and rapid analysis and detection Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1. Preparation of fluorescent biosensor and detection of miR34c
[0022] Target-assisted cyclic amplification process: Before the hairpin structure is used, hairpin 1 (HP1) and hairpin 2 (HP2) are heated in a water bath at 90 °C for 5 min to form a hairpin structure, and then slowly cooled to room temperature to form stem-loop DNA structure. Take 6 μL Tris buffer, 2 μL HP1 (10 μM), 2 μL HP2 (10 μM), 2 μL DP1 (10 μM) and 2 μL target (MicroRMA-34) (different concentrations), respectively, add 2 μL 10×phi29 polymerase Buffer, 2 μL 10× NEB Cutsmart Buffer, 0.5 μL phi29 (10,000 U / ml) polymerase, 0.5 μL Nt.BbvCI (10 U / μL) nickase and 1 μL dNTPs (10 mM) constitute a 20 μL cyclic amplification reaction system. The above reaction system was placed in a constant temperature oscillator at 37°C to react for 3.5h. Finally, the reaction was terminated by placing at 80 °C for 20 min to obtain S1.
[0023] Fluorescence quenching process: In order to achieve fluorescence quen...
Embodiment 2
[0025] Example 2. Preparation of fluorescent biosensor and detection of miR34c
[0026] Changed "First add 40 μL cCQD to 40nM FAM-probe fluorescent probe, incubate at 50°C for 30 min." to "First add 30 μL cCQD to 40nM FAM-probe fluorescent probe, incubate at 50°C 30min." Other preparation conditions were the same as in Example 1, and a biosensor with morphology and properties similar to those in Example 1 was obtained. The results of the detection of miR34c were the same as those in Example 1.
Embodiment 3
[0027] Example 3. Preparation of fluorescent biosensor and detection of miR34c
[0028] Will "take 6μL Tris buffer, 2μL HP1 (10μM), 2μL HP2 (10μM), 2μL DP1 (10μM) and 2μL target (MicroRMA-34) (different concentrations), add 2μL 10 × phi29 polymerase Buffer, 2μL 10×NEBCutsmart Buffer, 0.5μL phi29 (10,000U / ml) polymerase, 0.5μL Nt.BbvCI (10U / μL) nickase and 1μL dNTPs (10mM) constitute a 20μL cyclic amplification reaction system.” To "take 6 μL Tris buffer, 2 μL HP1 (10 μM), 2 μL HP2 (10 μM), 2 μL DP1 (10 μM), and 2 μL target (MicroRMA-34) (different concentrations), respectively, add 2 μL 10×phi29 polymerase Buffer, 2 μL 10×NEB Cutsmart Buffer, 0.4μL phi29 (10,000U / ml) polymerase, 0.4μL Nt.BbvCI (10U / μL) nickase and 1μL dNTPs (10mM) constitute a 20μL cyclic amplification reaction system.” Other preparation conditions were the same as those in Example 1, and a biosensor with morphology and properties similar to those in Example 1 was obtained. The results of the detection of mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com