Genetically engineered bacteria producing D-pantolactone hydrolase as well as construction method and application of genetically engineered bacteria
A technology of pantolactone and genetically engineered bacteria is applied in the field of construction of genetically engineered bacteria, which can solve the problems of low expression of D-pantolactone hydrolase and achieve the effect of efficient hydrolysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] A genetically engineered bacterium producing D-pantolactone hydrolase, the construction method comprising the steps of:
[0038] (1) Construction of cloning vector pMD19-T-DL
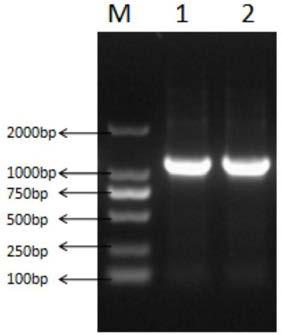
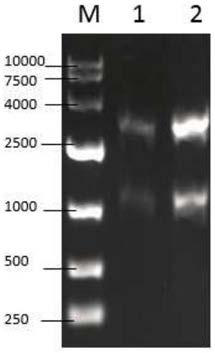
[0039] The cDNA sequence of D-pantolactone hydrolase (GenBank: AY728018.1) was found on NCBI, and sent to Suzhou Jinweizhi Biological Co., Ltd. for optimized synthesis according to the codons preferred by yeast to obtain optimized D-pantolactone hydrolase Acid lactone hydrolase gene (DL), the DL sequence is shown in SEQ ID NO:1. Using the synthetic gene (DL) as a template, PCR amplification was carried out with the designed primers, and the results were as follows: figure 1 shown. The amplified product was connected with pMD19-T-Simple Vector, the cloning vector pMD19-T-DL was constructed and transformed into E.coli JM109 competent, the positive transformant was screened by ampicillin resistance plate, and the recombinant plasmid was extracted by alkaline cleavage method. For specific methods, ...
Embodiment 2
[0055] Embodiment 2: the enzyme production culture of genetically engineered bacteria
[0056] Preparation of seed solution: Take 200 μL of the preserved bacterial solution in YPD liquid medium, cultivate at 30°C, 200 r / min for 36 hours, and use it as the primary seed. In a 250mL Erlenmeyer flask containing 50mL of YPD liquid medium, inoculate primary seeds at a transfer rate of 4% (volume ratio), and cultivate them at 30°C and 200r / min for 36h as secondary seeds.
[0057] Shake flask enzyme production culture: medium components are sucrose 40g / L, peptone 20g / L, beef extract 15g / L, NaCl 6g / L, KCl 6g / L, CaCl 2 3g / L, MgSO 4 3g / L, 30℃, 200r / min fermentation time 85h
[0058] Determination of biomass: use the dry weight of the bacteria contained in the fermentation broth per unit volume (L) for a certain period of time to represent the growth of the bacteria, separate the fermentation broth at 10000r / min for 5min to obtain the bacteria, and wash with sterile deionized water fo...
Embodiment 3
[0061] Embodiment 3: the application of genetically engineered bacteria
[0062] (1) Preparation of immobilized enzyme
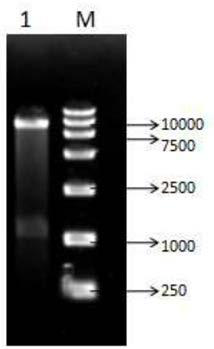
[0063] Put an appropriate amount of pretreated resin D380 into a 50mL Erlenmeyer flask, add a crude enzyme solution with a total enzyme activity of 12-80U per gram of resin, then place it in a shaker, and absorb it at 20-35°C at 100-200r / min for 2 ~6h, pre-cool at 4°C, then add glutaraldehyde to a final concentration of 0.1%~1%, crosslink for 0.5~3h, and finally wash repeatedly with deionized water to remove free enzyme. Or put an appropriate amount of pretreated resin D380 into a packed bed reactor such as Figure 6 As shown, use crude enzyme solution (total enzyme activity per gram of resin is 12-80U) for cyclic adsorption, 2-6h, pre-cool at 4°C, and then use 0.1%-1% glutaraldehyde for cyclic cross-linking for 0.5-3h, Finally, the free enzymes were removed by repeated washing with deionized water.
[0064] (2) Hydrolysis of DL-pantolactone by immobilize...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com