Chitin deacetylase derived from saccharomyces cerevisiae, encoding gene and applications
A technology of deacetylase and Saccharomyces cerevisiae, applied in the directions of application, genetic engineering, plant genetic improvement, etc., can solve the problems such as chitin deacetylase that have not been reported yet, and achieve the effect of a wide range of pH adaptation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Full-length cloning of chitin deacetylase gene.
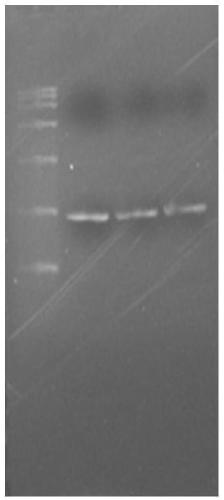
[0036] The target gene was upregulated from NCBI, and the target gene was synthesized in Zhongmei Taihe Biotechnology (Beijing) Co., Ltd. After performing multiple sequence alignment analysis on chitin deacetylases of the carbohydrate esterase family 4 (CE4), primers CDA-F: 5'-CCATGG GAAGCTAATAGGGAAGATTTA-3'; CDA-R5' were designed -CCGCTCGAGGGACAAGAATTCTTTTATGT AATC-3'. PCR amplification was performed using the synthesized DNA as a template. The PCR reaction conditions were: 94°C for 5 min, 1 cycle; 94°C for 30 s, 45°C for 30 s, 72°C for 1 min 30 s, 30 cycles; 72°C for 10 min, 1 cycle. After the PCR product was analyzed by agarose gel electrophoresis, the target fragment was recovered by cutting the gel, connected to the pMD19-T vector and then sequenced.
Embodiment 2
[0037] Embodiment 2 Chitin deacetylase gene complete sequence analysis
[0038] The sequencing results were analyzed using the Basic Local Alignment Search Tool (BLAST) in the GenBank database, and the Vector NTI Suite 8.0 software was used for multiple sequence alignments to analyze their homology. The domains of the sequences were analyzed using the SimpleModular Architecture Research Tool (SMART) online tool.
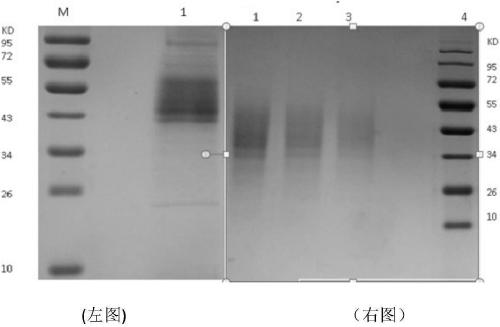
[0039] The obtained chitin deacetylase (named ScCDA 2 ) coding region is 861bp long, and its nucleotide sequence is shown in SEQ ID NO 1. The nucleotide sequence of ScCDA2 has the highest identity (100%) with the deacetylated protein gene (accession number CP020202.1) derived from Saccharomyces cerevisiae strainY12chromosome XII sequence. cCDA2 encodes 287 amino acids and a stop codon, its amino acid sequence is shown in SEQ ID NO 2, the protein molecular weight is 34kDa, and the predicted isoelectric point is 7.0. SMART analysis showed that the protein domain cha...
Embodiment 3
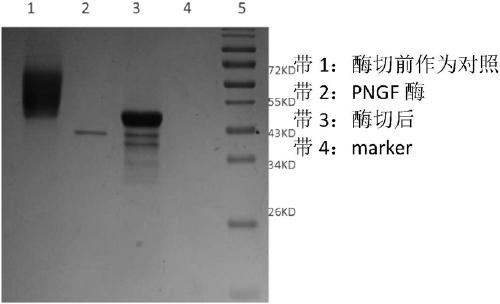
[0040] Example 3 ScCDA 2 The recombinant expression of the gene in Pichia pastoris uses the synthesized DNA as a template, and uses the designed primers CDA-F: 5'-CCATGGGAAGCTAATAGGGAAGATTTA-3'; CDA-R5'-CCGCTCGAGGGACAAGAATTCTTTTATGTAATC-3' to amplify the target gene sequence. Construct the target gene on the PMD-19T cloning vector, spread it on the solid plate of Luria-Bertani medium containing 100 μg / mL ampicillin, culture at 37°C for 14 hours, and pick a single clone; Ampicillin was cultured in liquid Luria-Bertani medium, and the plasmid was extracted; the plasmid was verified by colony PCR with the forward primer ScCDA-F and the reverse primer ScCDA-R, and the result was an amplified product of the correct size, which preliminarily proved that the constructed recombinant The plasmid is correct; then the recombinant plasmid is sent to Yingwei Jieji Company for sequencing, and the results show that the ScCDA shown in SEQ ID NO 1 is inserted between the Nco I and Xho I sites ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com