Method for evaluating CRISPR/Cas9 gene editing efficiency or miss frequency
A gene editing, off-target technology, applied in the field of bioengineering, can solve the problems of time-consuming, labor-intensive, low sensitivity, and inapplicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
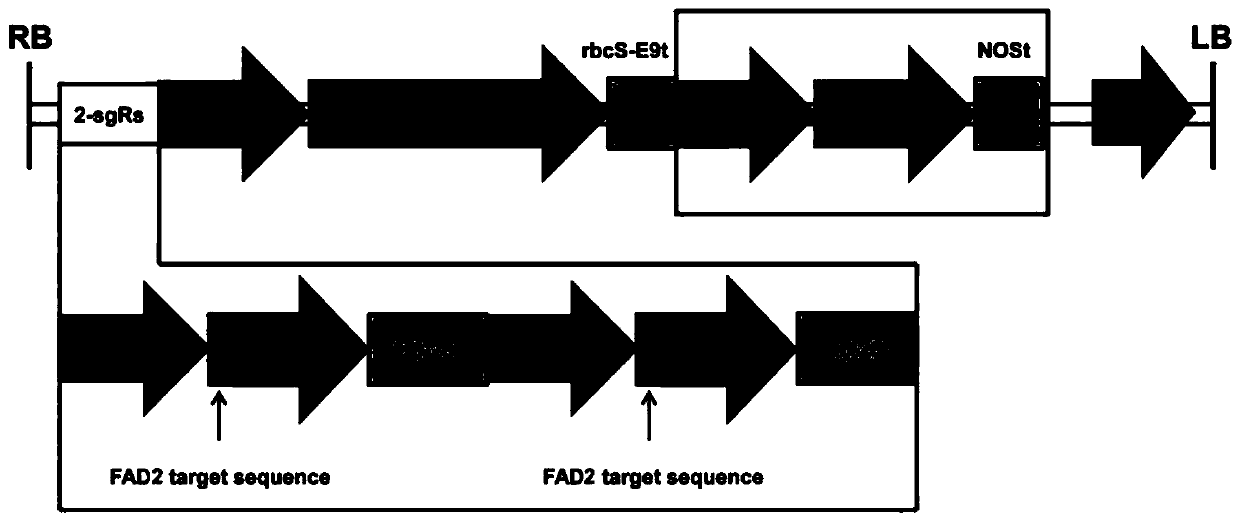
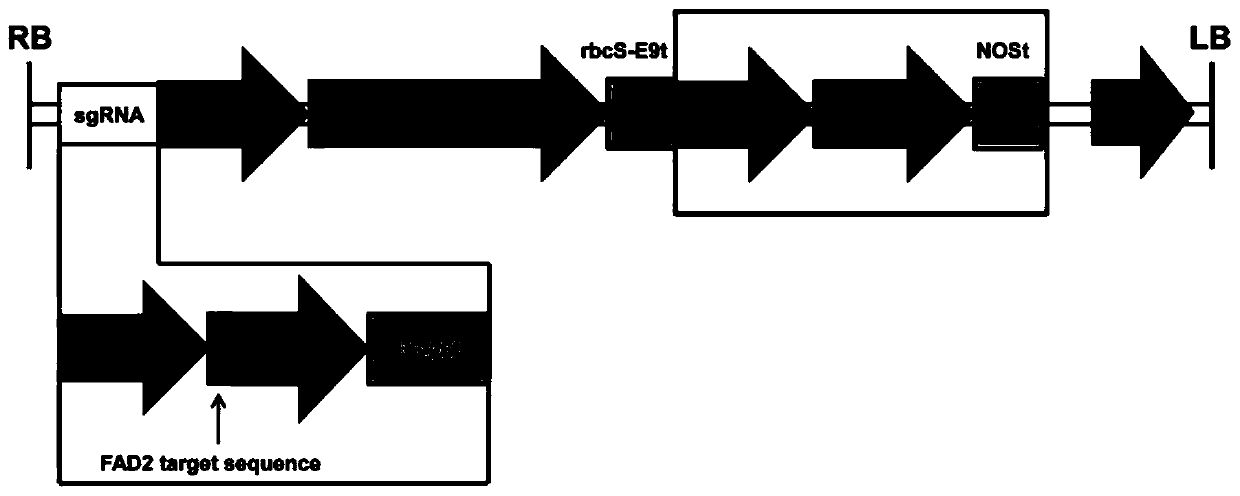
[0045] 1. Construction of CRISPR / Cas9 vector
[0046] 1.1 Cloning the reporter gene into the CRISPR / Cas9 vector
[0047] Using Arabidopsis genomic DNA as a template, ZYP11-FP (such as SEQ ID NO: 17) and ZYP11-BP (such as SEQ ID NO: 18) as primers, amplify the seed-specific expression of the At2S3 gene promoter to drive mCherry fluorescent protein gene; after electrophoresis detection has the At2S3 band, the first PCR product is diluted 100 times, and the PCR product after dilution is used as a template, ZYP12-FP (such as SEQ ID NO: 19) and ZYP12-RP (such as SEQ ID NO: 19) and ZYP12-RP (such as SEQ ID NO: 19) ID NO: 20) was used as the primer for the second amplification, and the restriction sites BamHI and EcoRI were cloned to both ends of the At2S3 fragment. The PCR product was BamHI-EcoRI-At2S3-BamHI, and its length was 455bp. The amplified The product was recovered from the gel for future use.
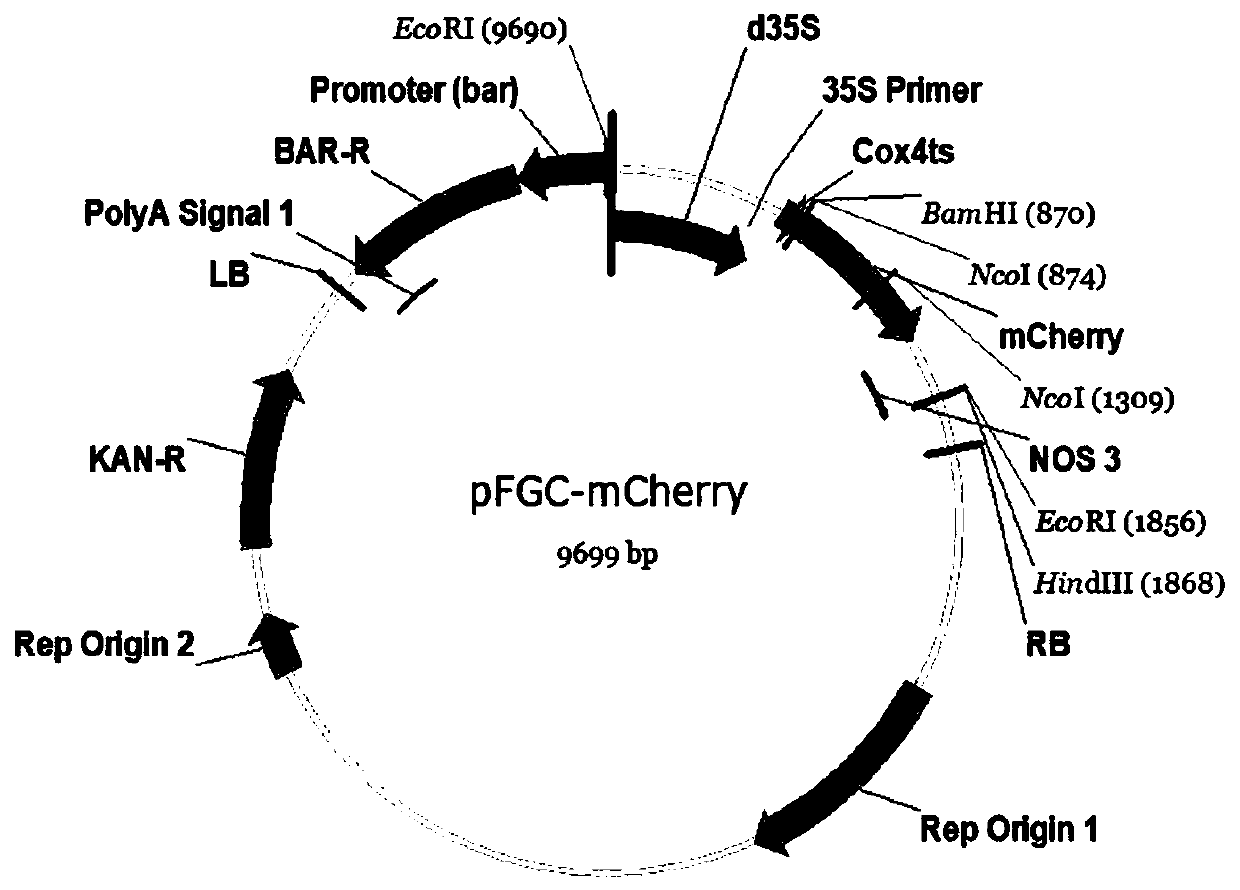
[0048] Digest the pFGC-35S-mCherry-NOS plasmid vector with BamHI (plasmid map...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com