Specific primer, kit and method for qualitatively and/or quantitatively detecting lactobacillus rhamnosus
A technology for quantitative detection of Lactobacillus rhamnosus, applied in the field of genetic engineering, to achieve high sensitivity, fast and accurate qualitative, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Metagenomic DNA Extraction from Fecal Samples
[0055] Take 0.5g of the sample and place it in a 2mL centrifuge tube, add 1000μL Inhibit EX lyase, shake for 10min; put it in an 80°C water bath for 10min after shaking, vortex for 15s, and centrifuge at 12000×g for 5min at room temperature; collect the supernatant and transfer to another Add 25 μL proK to a centrifuge tube, preheat AL buffer 300 μL at 70°C, shake for 5 minutes; bathe in water at 70°C for 10 minutes; add 300 μL of absolute ethanol, absorb all the liquid to the spin column after shaking, centrifuge at 12000×g for 2 minutes; add 500 μL AW1, let stand 1min, centrifuge at 12000×g for 2min; add 500μLAW2, let it stand for 1min, centrifuge at 12000×g for 2min; spin for 2min, change the collection tube and redissolve for later use. The DNA extraction kit is QIAamp FastDNA Stool Mini Kit (50), Cat.No.51604.
Embodiment 2
[0057] Qualitative detection of specific primers for Lactobacillus rhamnosus M9
[0058] Based on a specific fragment (SEQ ID NO.3) in the whole genome sequence of Lactobacillus rhamnosus M9, the following primer sequences were designed:
[0059] Upstream primer M9F: 5'-GTAATGTAAATGGGGTTCCTGTG-3' (SEQ ID NO.1);
[0060] Downstream primer M9R: 5'-TGGTTTCCCCTATAATCGTTGTCC-3' (SEQ ID NO.2).
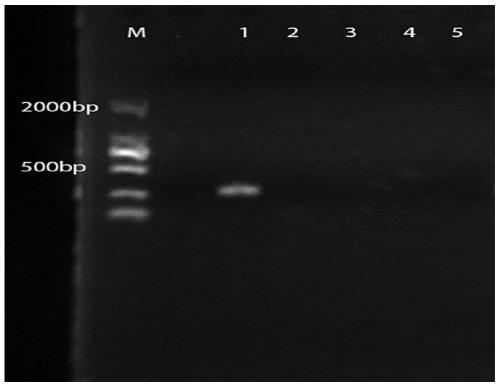
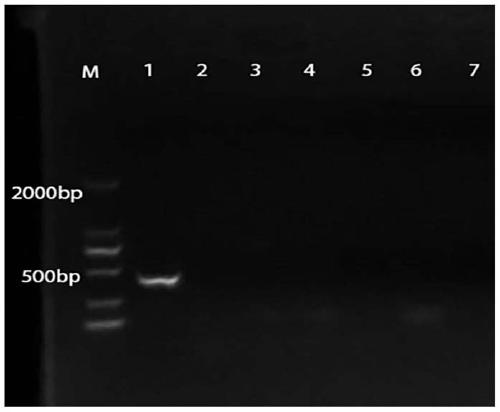
[0061] After designing the specific primers for Lactobacillus rhamnosus M9, the specificity of the primers was verified, and four strains of rhamnosus with higher homology to Lactobacillus rhamnosus M9 in taxonomic status were selected. Lactobacillus was amplified by PCR as a control.
[0062] A total of 12 strains of lactic acid bacteria were used in this test, including 5 strains of Lactobacillus rhamnosus including M9, 1 strain of Lactobacillus casei and 6 strains of Lactobacillus plantarum. The above strains were obtained from the Lactic Acid Bacteria Resource Bank (LACBB) of Inner Mo...
Embodiment 3
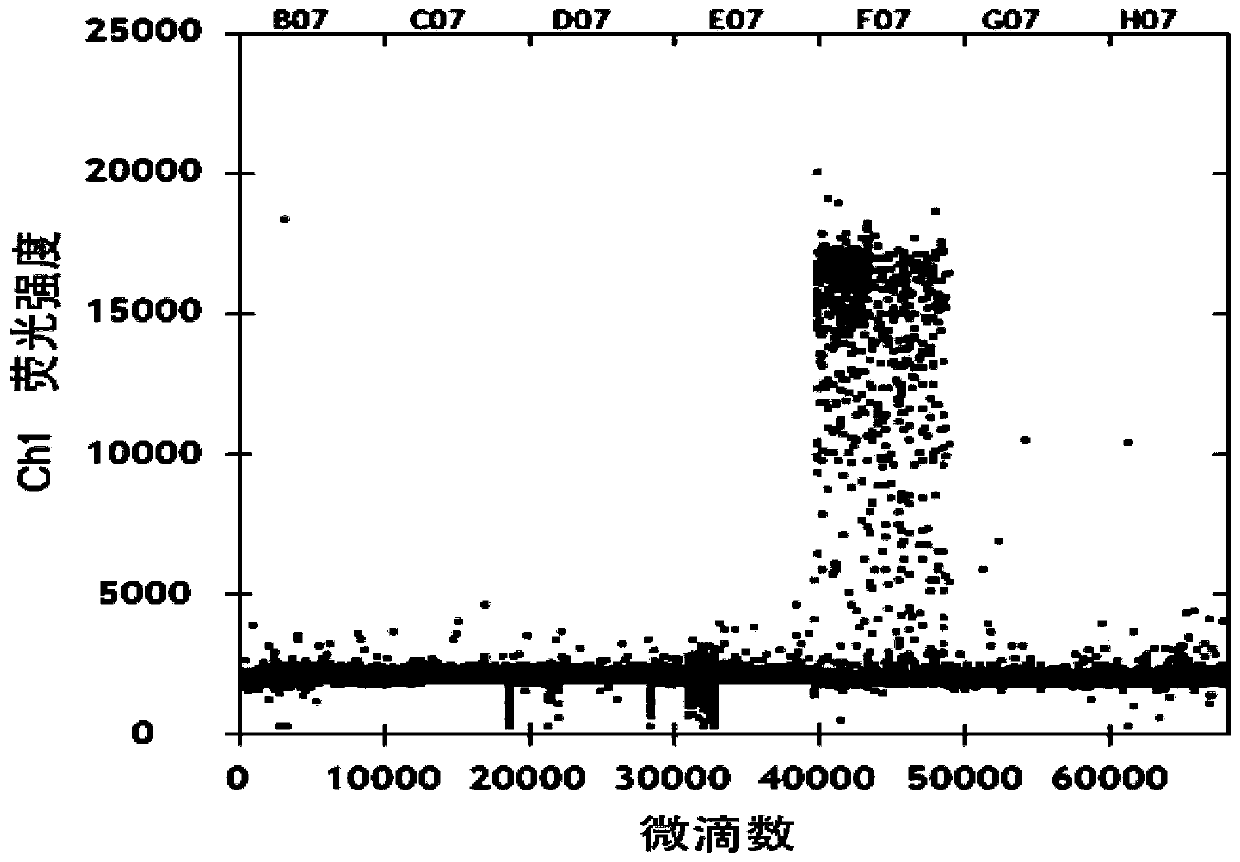
[0073] Quantitative detection of specific primers for Lactobacillus rhamnosus M9
[0074] Amplification system: 20μL: 10×PCR supermix 10μL, 10mol / L upstream and downstream primers 0.2μL each, DNA template 10ng 2μL, ddH 2 O supplemented to 20 μL;
[0075] The amplification conditions were: denaturation at 95°C for 10 min; denaturation at 94°C for 30 s, annealing at 54°C for 1 min, and 30 cycles; extension at 4°C for 5 min, extension at 90°C for 5 min, and storage at 4°C.
[0076] Put the PCR-amplified droplets into the droplet analyzer. After the droplet analyzer absorbs all the droplets, they are divided into positive droplets and negative droplets according to the fluorescence intensity; among them, the positive droplets are Lactobacillus rhamnosus The amplified fragment of M9 strain DNA, the negative micro-droplet is non-M9 DNA fragment, according to the percentage of positive micro-droplets in the total micro-droplets, according to the formula (copy number × 20) ÷ 2 × retu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com