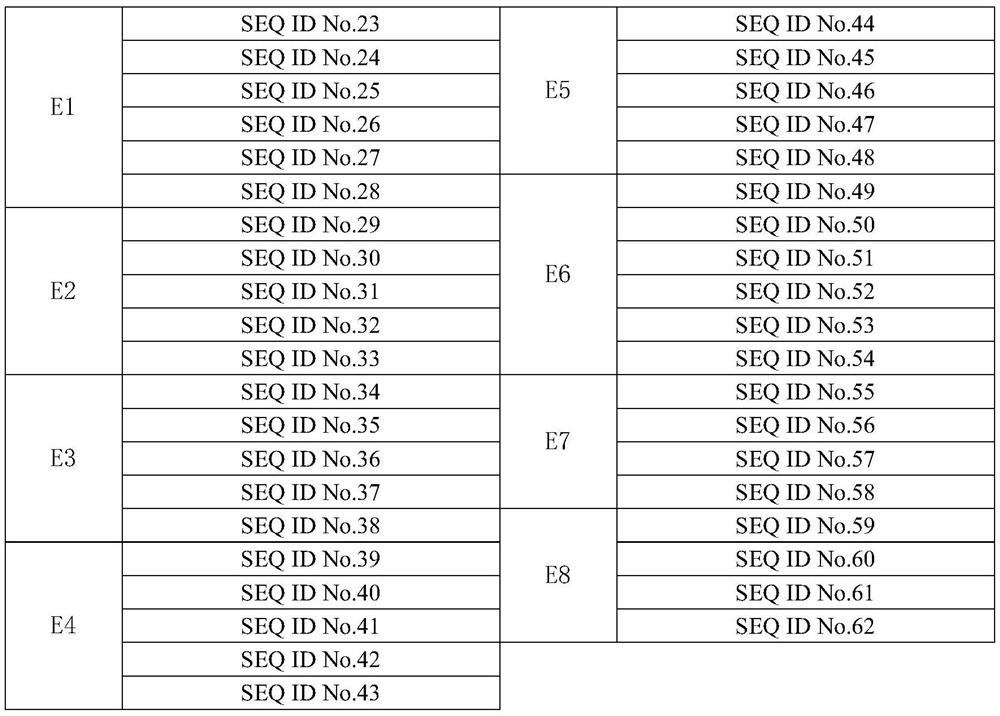
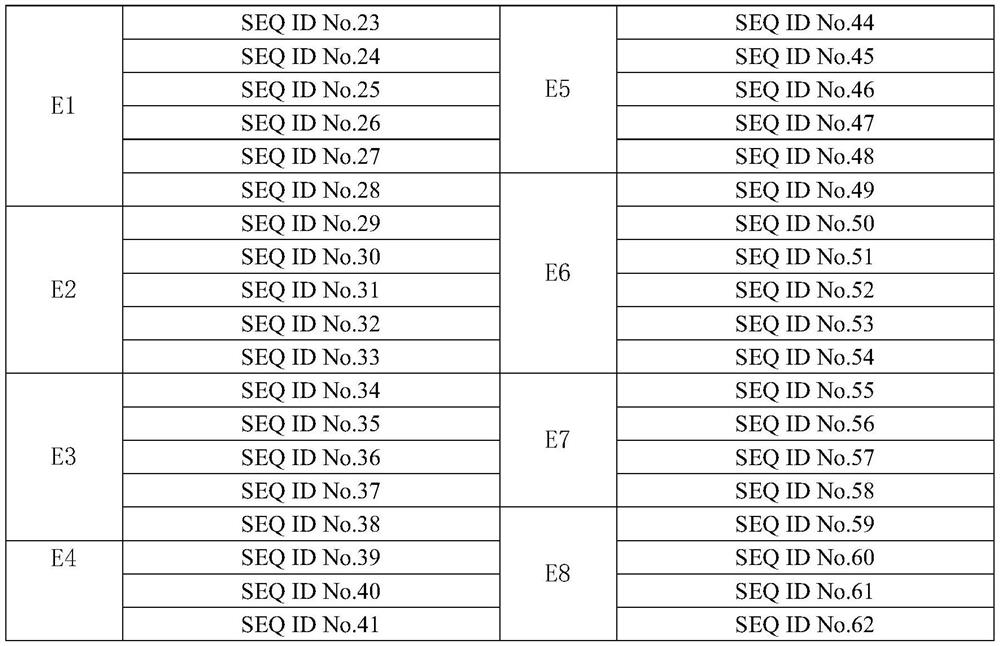
Primer set, kit and method for detecting lung cancer-related gene mutation in human circulating tumor DNA
A kit and tumor technology, applied in recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of low efficiency of clinical cancer treatment, reduce the burden of medical treatment, low cost, and sample input. less effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1 Detection of lung cancer-related gene mutation sites by the technology of the present invention
[0073] 1. Primer design and synthesis
[0074]针对BRAF_p.D594G、BRAF_p.G469A、BRAF_p.G469V、BRAF_p.V600E、EGFR_p.C797S、EGFR_p.D770_N771insSVD、EGFR_p.D770_N771insG、EGFR_p.E709A、EGFR_p.E709G、EGFR_p.E709K、EGFR_p.E746_A750del、EGFR_p.E746_E749del、EGFR_p .E746_S752>D、EGFR_p.E746_S752>V、EGFR_p.E746_T751del、EGFR_p.G719A、EGFR_p.G719C、EGFR_p.G719S、EGFR_p.H773_V774insNPH、EGFR_p.H773_V774insH、EGFR_p.L747_A750>P、EGFR_p.L747_E749del、EGFR_p.L747_P753>S 、EGFR_p.L858R、EGFR_p.L861Q、EGFR_p.L861R、EGFR_p.S768I、EGFR_p.T790M、EGFR_p.V769_D770insASV、ERBB2_p.A775_G776insYVMA、ERBB2_p.G776>VC、KRAS_p.G12A、KRAS_p.G12V、KRAS_p.G12D、KRAS_p.G12R , KRAS_p.G12C, KRAS_p.G12S, KRAS_p.G13C, KRAS_p.G13D, KRAS_p.Q61H, KRAS_p.Q61K, KRAS_p.Q61E, KRAS_p.Q61P, KRAS_p.Q61L, KRAS_p.Q61R, PIK3CA_p.E542K, PIK .H1047L, PIK3CA_p.H1047R. Wait for 49 gene mutation sites used to assess the specificity and sensitivity of...
Embodiment 2
[0147] Embodiment two: standard product detection
[0148] 9 cases of standard DNA samples (purchased from Horizon Company) with known mutations were selected for detection, numbered A1-A9. Among them, the mutation frequency of each mutation site corresponding to each sample of A1-A8 is 0.1%, and A9 is wild type.
[0149] According to the method of the present invention, according to the steps described in Example 1, 9 cases of standard DNA samples were detected by mass spectrometry. According to the judgment standard described in the embodiment, the results of the 9 standard samples were analyzed, and Table 14 was obtained.
[0150] Table 14
[0151]
[0152]
[0153] As a result of the test, the mutations corresponding to the 8 mutation-type standard samples can be detected normally, which shows that the method of the present invention can detect the sites with a mutation frequency as low as 0.1% of the 20ng sample amount, and has frontier advantages.
Embodiment 3
[0154] Example 3: Detection of free DNA samples
[0155] According to the method of the present invention, according to the steps described in Example 1, mass spectrometry was performed on the cell-free DNA samples of 30 lung cancer patients, and the sample numbers were B1-B30. The results of the 30 samples were analyzed according to a criterion described in the embodiment, and Table 15 was obtained.
[0156] Table 15
[0157] sample detected mutation sample detected mutation B1 KRAS_p.G12D B16 EGFR_p.S768I B2 KRAS_p.G12C B17 EGFR_p.L858R B3 EGFR_p.G719S, EGFR_p.L861Q B18 ERBB2_p.A775_G776insYVMA B4 EGFR_p.G719S, EGFR_p.L861Q B19 KRAS_p.G12S B5 EGFR_p.T790M B20 EGFR_p.E746_A750del B6 EGFR_p.E746_A750del B21 KRAS_p.G12D B7 BRAF_p.V600E B22 EGFR_p.V769_D770insASV B8 EGFR_p.S768I, EGFR_p.L858R B23 EGFR_p.E746_A750del B9 EGFR_p.L861Q B24 EGFR_p.S768I, EGFR_p.L858R B10 EGFR_p.L858...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com