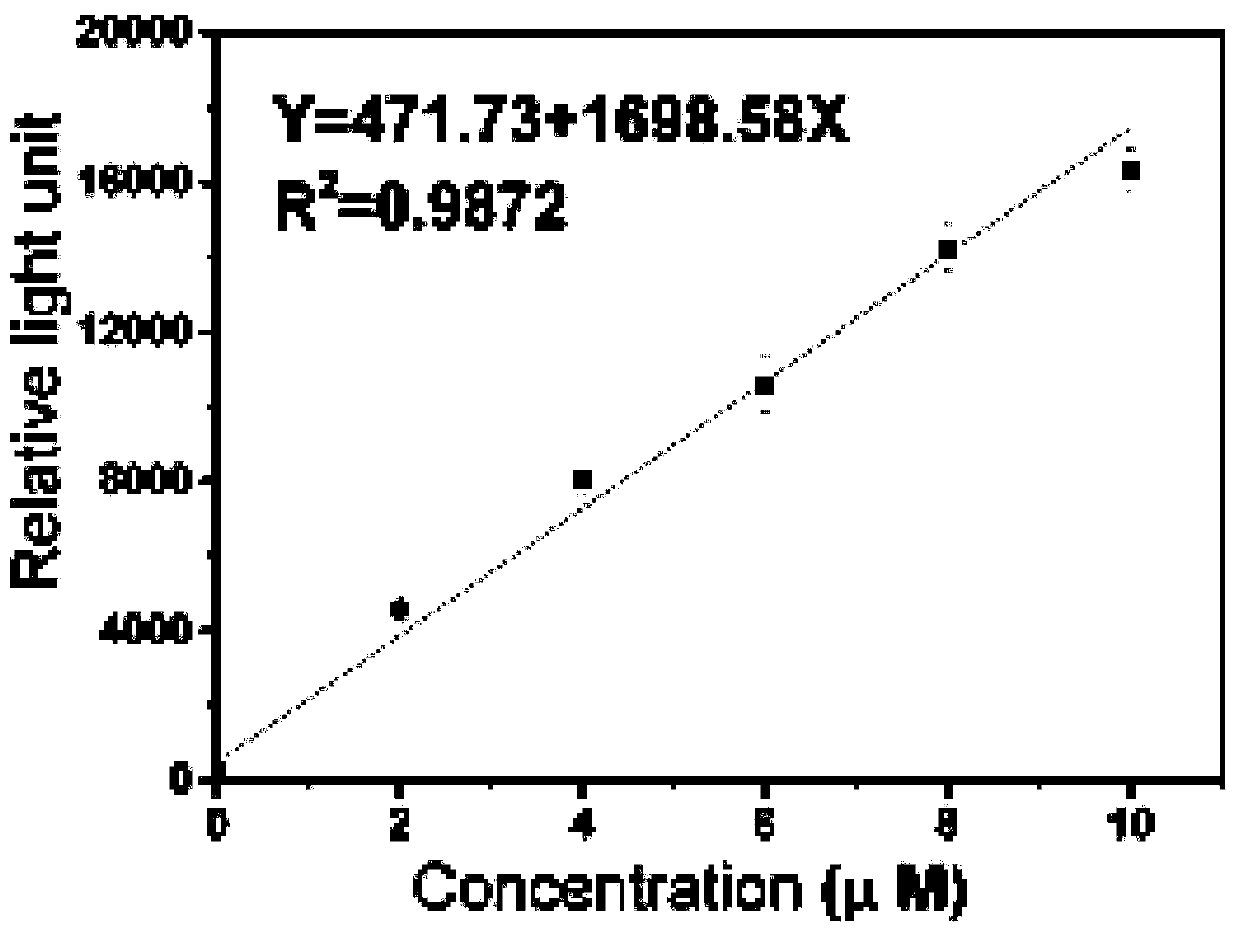
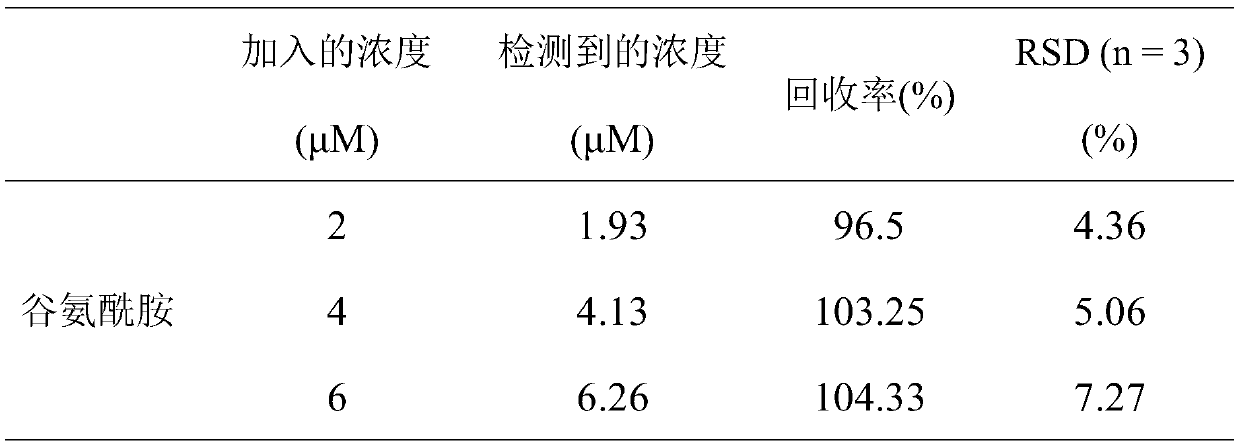
Glutamine detection method based on double enzyme coupling
A technology of glutamine and detection method, applied in the field of glutamine determination, can solve problems such as damage to plant samples, lack of a specific detection method for glutamine, etc., and achieve the effects of convenient operation, low price and little damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038](1) Prepare the TE buffer solution (10mM tris-hydrochloric acid, 1mM sodium edetate, pH 7.4) for dissolving the required DNA, respectively, the sequences are 5'-TTTGGGTAGGGCGGGTTGGG-3', 5 The DNA of the two segments of DNA of '-CGAACTCTGCAACATAAAAAA-3' was dissolved in the above-mentioned TE buffer solution to obtain DNA solution A and DNA solution B with a concentration of 10 μM, and stored at 4°C;
[0039] (2) prepare potassium phosphate buffer solution (200mM K 2 HPO 4 -KH 2 PO 4 , 400mM NaCl, pH 7.4), using a small amount of dimethyl sulfoxide to dissolve hemin, and then prepare with the potassium phosphate buffer to obtain a heme solution with a concentration of 10 μM, and store it at 4°C;
[0040] (3) Mix the above DNA solution A, DNA solution B and heme solution at a volume ratio of 1:1:2, let stand at room temperature for 30-60 minutes to obtain a DNA mimic enzyme solution, and store at 4°C;
[0041] (4) Prepare glutamine synthetase working solution, the reac...
Embodiment 2
[0047] (1) Prepare the TE buffer solution (10mM tris-hydrochloric acid, 1mM sodium edetate, pH 7.4) for dissolving the required DNA, respectively, the sequences are 5'-TTTGGGTAGGGCGGGTTGGG-3', 5 The DNA of the two segments of DNA of '-CGAACTCTGCAACATAAAAAA-3' was dissolved in the above-mentioned TE buffer solution to obtain DNA solution A and DNA solution B with a concentration of 10 μM, and stored at 4°C;
[0048] (2) prepare potassium phosphate buffer solution (200mM K 2 HPO 4 -KH 2 PO 4 , 400mM NaCl, pH 7.4), using a small amount of dimethyl sulfoxide to dissolve hemin, and then prepare with the potassium phosphate buffer to obtain a heme solution with a concentration of 10 μM, and store it at 4°C;
[0049] (3) Mix the above DNA solution A, DNA solution B and heme solution at a volume ratio of 1:1:2, let stand at room temperature for 30-60 minutes to obtain a DNA mimic enzyme solution, and store at 4°C;
[0050] (4) prepare glutamine synthetase working solution, this re...
Embodiment 3
[0056] (1) Prepare the TE buffer solution (10mM tris-hydrochloric acid, 1mM sodium edetate, pH 7.4) for dissolving the required DNA, respectively, the sequences are 5'-TTTGGGTAGGGCGGGTTGGG-3', 5 The DNA of the two segments of DNA of '-CGAACTCTGCAACATAAAAAA-3' was dissolved in the above-mentioned TE buffer solution to obtain DNA solution A and DNA solution B with a concentration of 10 μM, and stored at 4°C;
[0057] (2) prepare potassium phosphate buffer solution (200mM K 2 HPO 4 -KH 2 PO 4 , 400mM NaCl, pH 7.4), using a small amount of dimethyl sulfoxide to dissolve hemin, and then prepare with the potassium phosphate buffer to obtain a heme solution with a concentration of 10 μM, and store it at 4°C;
[0058] (3) Mix the above DNA solution A, DNA solution B and heme solution at a volume ratio of 1:1:2, let stand at room temperature for 30-60 minutes to obtain a DNA mimic enzyme solution, and store at 4°C;
[0059] (4) Prepare glutamine synthetase working solution, the rea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com