PCR primers and method for detecting composition of environmental microorganism arsenic oxidation gene species
A technology for detecting environment and arsenic oxidation, which is applied in the field of biotechnology detection to achieve efficient analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The design of aioA gene PCR primers comprises the following steps:
[0028] (1) From KEGG (https: / / www.kegg.jp / ) and NCBI Refseq ( https: / / www.ncbi.nlm.nih.gov / refseq / ) database to download the full-length aioA protein sequence and the corresponding gene sequence;
[0029] (2) Muscle software (http: / / www.drive5.com / muscle) compared the aioA protein sequence and gene sequence to determine the gene sequence of the molybdenum coenzyme conserved domain.
[0030] (3) With the gene sequence encoding the conserved domain of the molybdenum coenzyme as the target, apply the DegePrime software ( https: / / github.com / EnvGen / DegePrime ) to design primers.
[0031] (4) According to the results of PrimerMatching and PrimerDeg of DegePrime software, select optimized primers, select forward primer (aioA-1109F1): 5'-atctggggbaayracaayta-3' (SEQ ID NO.1), reverse primer (aioA-1548R1) : 5'-ttcatbgasgtsagrttcat-3' (SEQ ID NO. 2).
Embodiment 2
[0033] Verification of the generality of the designed aioA primers by bioinformatics:
[0034] Align the primers designed in Example 1 to the full-length aioA gene sequence, and compare the effectiveness of the primers to the aioA genes of different species, the steps are as follows:
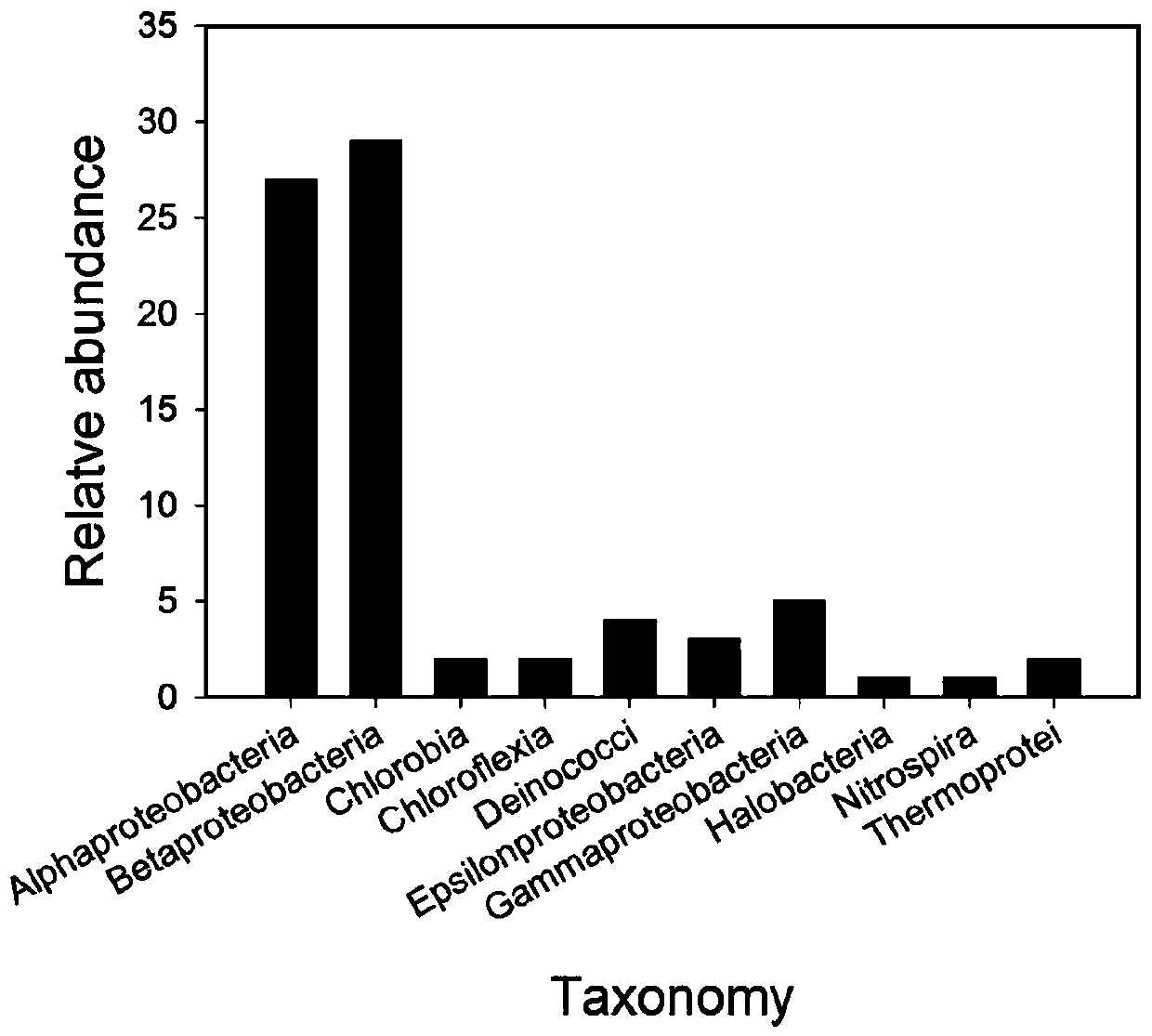
[0035] (1) from KEGG ( https: / / www.kegg.jp / ) and NCBI Refseq ( https: / / www.ncbi.nlm.nih.gov / refseq / ) database downloaded the full-length aioA gene sequence, and its species composition is shown in Table 1 and figure 1 .
[0036] Table 1. Species composition of the full-length aioA gene at the genus level
[0037]
[0038]
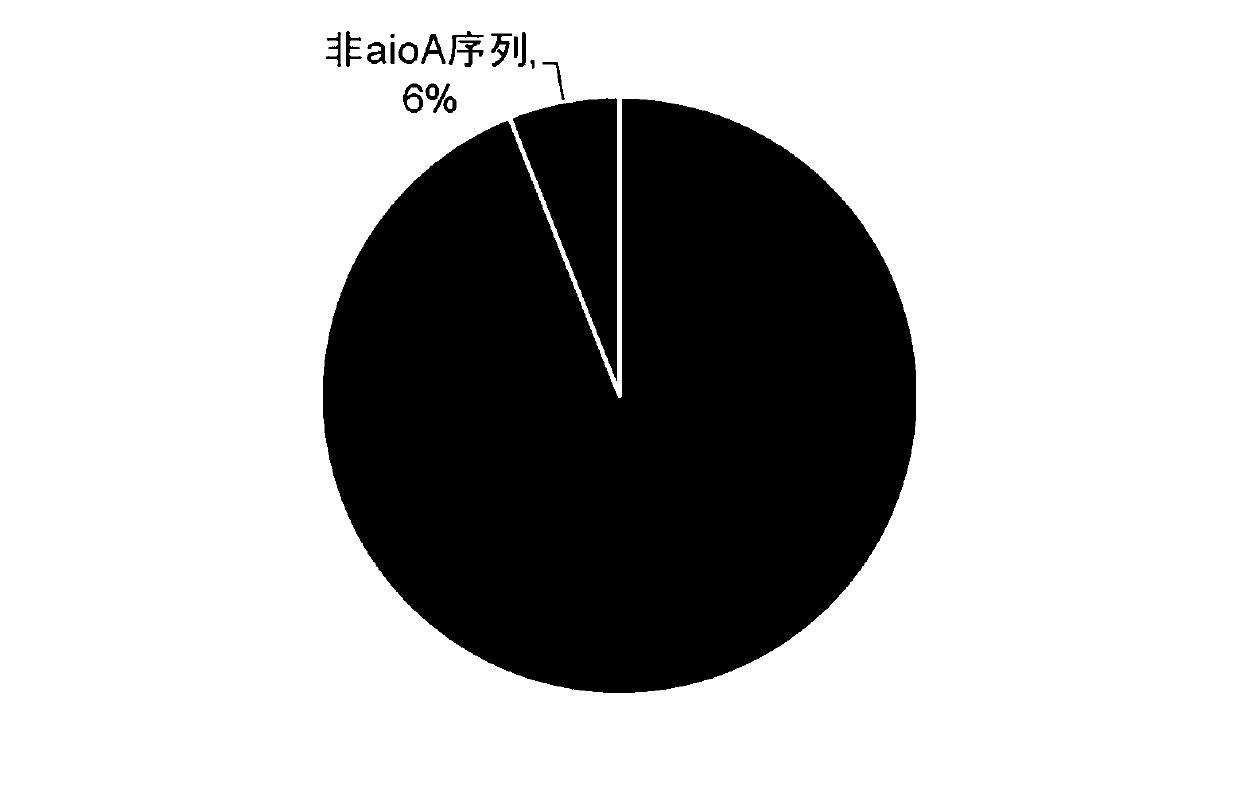
[0039] (2) The primer sequences were aligned to 79 full-length aioA gene sequences using Blastn software, and the alignment standard was 2 maximum mismatched bases, and the alignment similarity was greater than 90%. The ratio of aioA-62F1+aioA-518R1 to the full-length aioA gene sequence was 81.0%, and the comparison to the full-length aioA is shown in Table 2...
Embodiment 3
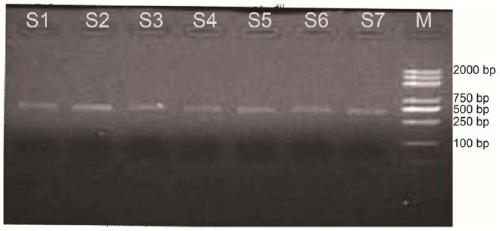
[0046]Extract paddy field soil microbial genomic DNA, PCR amplify aioA gene, a total of 7 paddy field soil samples, the steps are as follows:
[0047] (1) Weigh 0.25g fresh paddy field soil sample, use Powersoil TM DNA extraction kit (MOBIO company) was used to extract soil DNA samples. The quality of the extracted DNA samples was determined by horizontal electrophoresis and the concentration was determined by Nanodrop.
[0048] (2) Take 1 μl of environmental DNA as a template to amplify the aioA gene. aioA-1109F1+aioA-1548R1 were used as primers for PCR amplification. The total volume of the PCR amplification system is 50 μl, and the composition of the reaction system is: 1 μl of upstream and downstream primers (concentration 10M), 2 μl of 10×ExTag reaction solution (Shanghai Dalian Bao Biological Company), 1 μl (0.5U) of ExTag enzyme (Shanghai Dalian Bao Biological company), 2μl dNTPs (2.5mM), and 5μl sterile water. The PCR program was pre-denaturation at 94°C for 5 min...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com