SNP molecular marker related to lean meat rate, eye muscle area and eye muscle thickness on swine chromosome 6 and application thereof
An eye muscle area and molecular marker technology, applied in the field of molecular biotechnology and molecular markers, can solve the problems of restricting the application of molecular markers for complex traits and large confidence regions, and achieve increasing core competitiveness, increasing eye muscle area, and speeding up breeding. effect of the process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] (1) Experimental animals
[0037] The group of experimental pigs used in the present invention is 3916 purebred American Durocs from the Breeding Pig Branch of Wen's Food Group Co., Ltd., which is the core group of the Breeding Pig Branch.
[0038] In this experiment, Chinese and American Duroc pigs were selected from this resource group. The pigs have free access to food and water, and the entire feeding method and feeding conditions are always consistent, which is a conventional method.
[0039] (2) Sample collection
[0040] The above-mentioned piglet docked tail and ear tissues were collected and soaked in ethanol solution with a volume fraction of 75%, and stored in a -20°C refrigerator for later use.
[0041] (3) 50K SNP typing in the whole pig genome
[0042] The ear tissue or docked tail tissue collected from each individual of the 3916 Duroc breeding pigs selected from the above-mentioned resource groups, the whole genome DNA was extracted by the standard phe...
Embodiment 2
[0059] Example 2 Target DNA sequence amplification and sequencing
[0060] (1) Primer design
[0061] The DNA sequence of SEQ ID NO: 1 on pig chromosome 6 was downloaded from Ensembl website (http: / / asia.ensembl.org / index.html). And use primer design software primer premier 6.0 to design primers. The DNA sequences of the designed primers are as follows:
[0062] P001-F: 5'-ACTCTTGCCTCCGACTTCTC-3',
[0063] P002-R: 5'-AGACCTGGTGACATAGTTGATGA-3';
[0064] (2) PCR amplification
[0065] Add 1 μL of DNA template, 3.4 μL of double distilled water, 5 μL of 2×Tag PCR StanMix with Loading Dye, and 0.3 μL of primers P001-F and P002-R into a 10 μL reaction system. The PCR reaction conditions were as follows: 5 minutes of pre-denaturation at 94°C, 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, and finally 5 min at 72°C.
[0066] (3) DNA sequence determination
[0067] DNA sequence sequencing identification: carried out in Shenzh...
Embodiment 3
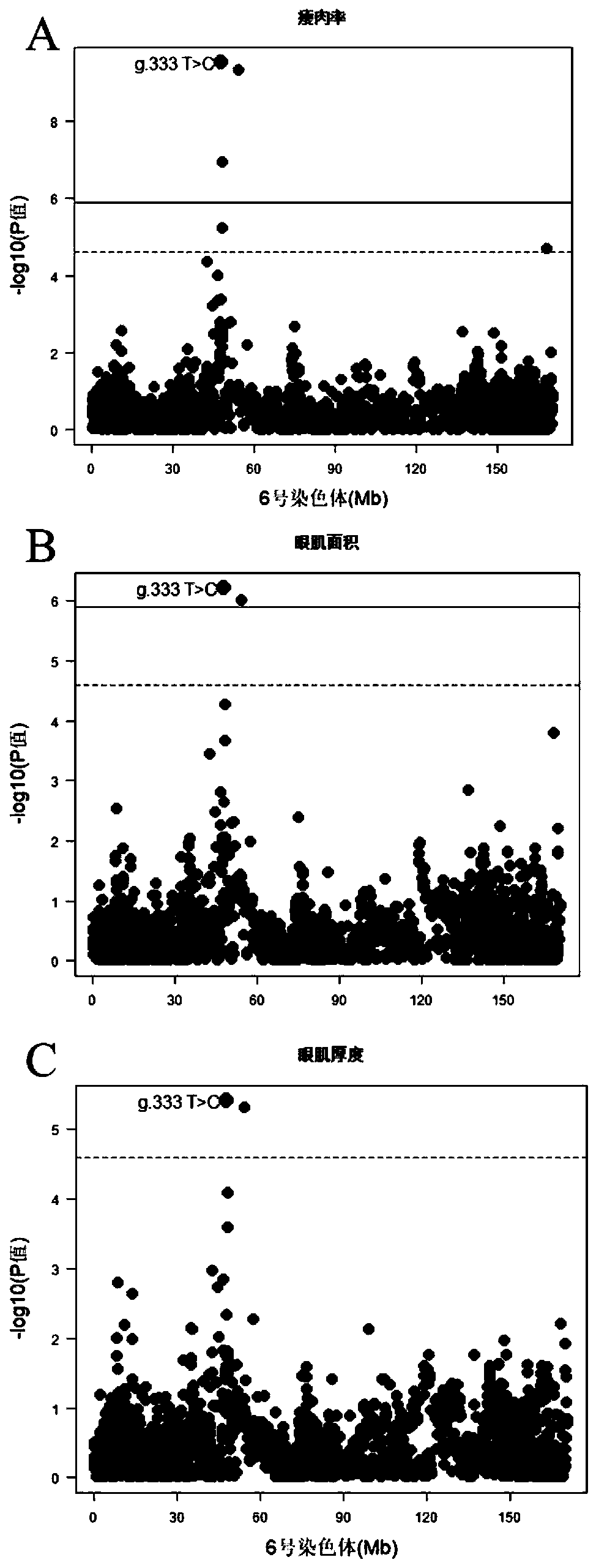
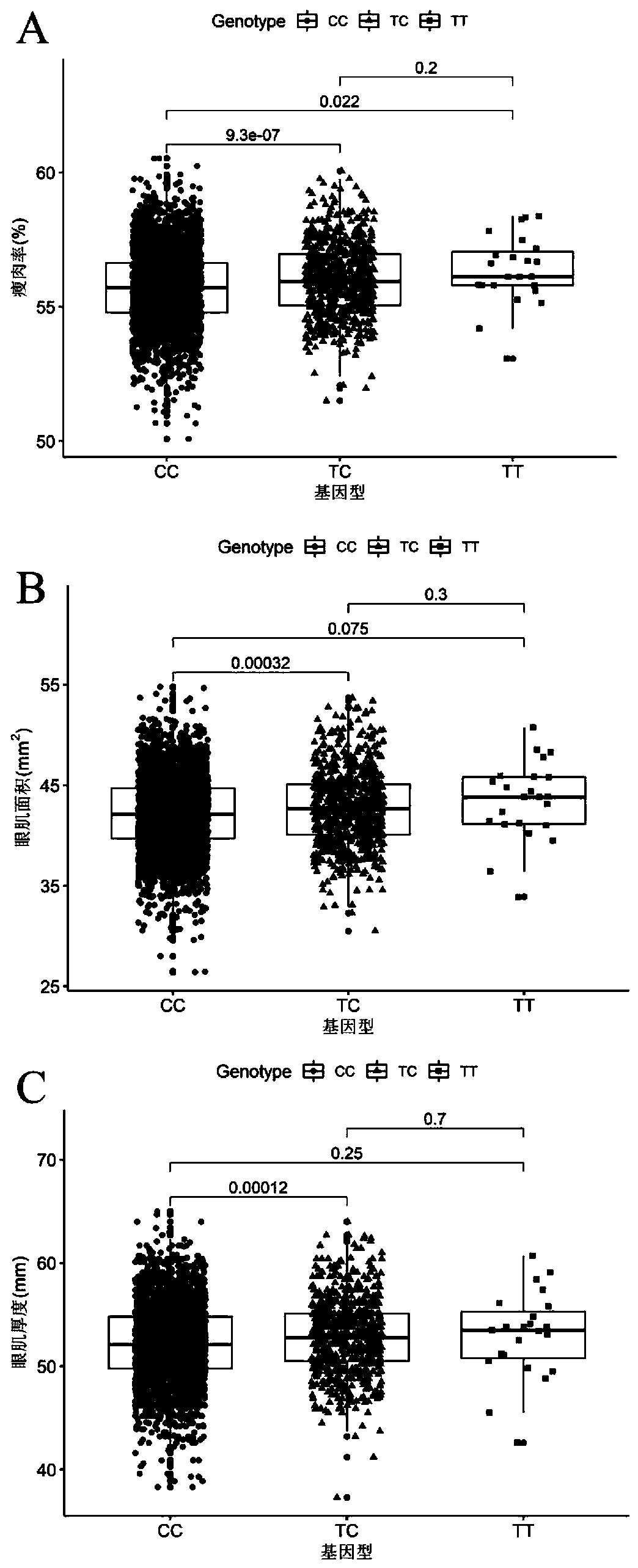
[0070] Example 3 SNP site g.333 T>C effect analysis of molecular markers
[0071] According to Tables 1 to 3, it can be seen that for lean meat percentage, eye muscle area and eye muscle thickness, the effect of SNP locus g. 1.28 and 0.76. Therefore, through molecular marker-assisted selection and gradually eliminating pigs with genotype CC in the population, the allele frequency of the allele T can be significantly increased, and the lean meat percentage, eye muscle area, and eye muscle thickness of the population can be improved. Pig growth and production performance are improved, and more high-quality commercial lean pork can be produced to meet people's demand for high-quality pork, thereby driving the growth of pork sales, which will bring huge economic benefits to enterprises.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com