Rapid detection and monitoring method for common swine infectious disease pathogens
A technology for infectious diseases and pathogens, applied in the field of molecular biology, to achieve the effects of good detection specificity, good specificity and compatibility, and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Discovery of nucleic acid detection sites for porcine infectious disease pathogens based on the CRISPR / Cas12a system
[0032]We obtained the genome sequence of the porcine infectious disease pathogen, and compared it through bioinformatics analysis to find the specific identification region of each strain of the porcine infectious disease pathogen. The swine infectious disease pathogens we screened in this patent are: African swine fever virus (ASFV), porcine foot and mouth disease virus (Foot and Mouth Disease virus, FMDV), porcine epidemic diarrhea virus (Porcine epidemic diarrhea virus, PEDV), porcine rotavirus group A (Porcine rotavirus group A, PRV), porcine transmissible gastroenteritis virus (Porcine transmissible gastroenteritis virus, TGEV).
[0033] The specific operation method is as follows: search for the whole genome sequence of the pathogen in the NCBI nucleic acid sequence library, obtain all the existing whole genome sequences of the pathogen,...
Embodiment 2
[0038] Example 2 Case of detecting nucleic acid of porcine infectious disease pathogen based on CRISPR / Cas12a system
[0039] 1. CRISPR / Cas12a gene cloning and protein expression
[0040] The Cas12a protein gene derived from Lachnospiraceae bacterium is used, and the codon is optimized to make the gene more suitable for expression in mammalian cells. The optimized Cas12a protein gene was cloned into the pET28a plasmid with a 6-His histidine tag to facilitate protein purification and expression. The Cas12a protein recombinant expression vector was transformed, and the expression strain was BL21 star (DE3).
[0041] The specific protein expression conditions are: in culture solution OD 600 When =0.6, add 0.5mMIPTG and cultivate for 4 hours. Bacteria were collected for protein purification. The purification conditions are: resuspend the bacteria in the lysate (50mM Tris, pH8.0, 300mM NaCl, 5% glycerol, 20mM imidazole), and perform sonication (70% amplitude, 2s On / 4s Off, 3 mi...
Embodiment 3
[0070] Example 3 Detection method of multiple porcine infectious disease pathogen nucleic acid based on CRISPR / Cas12a system
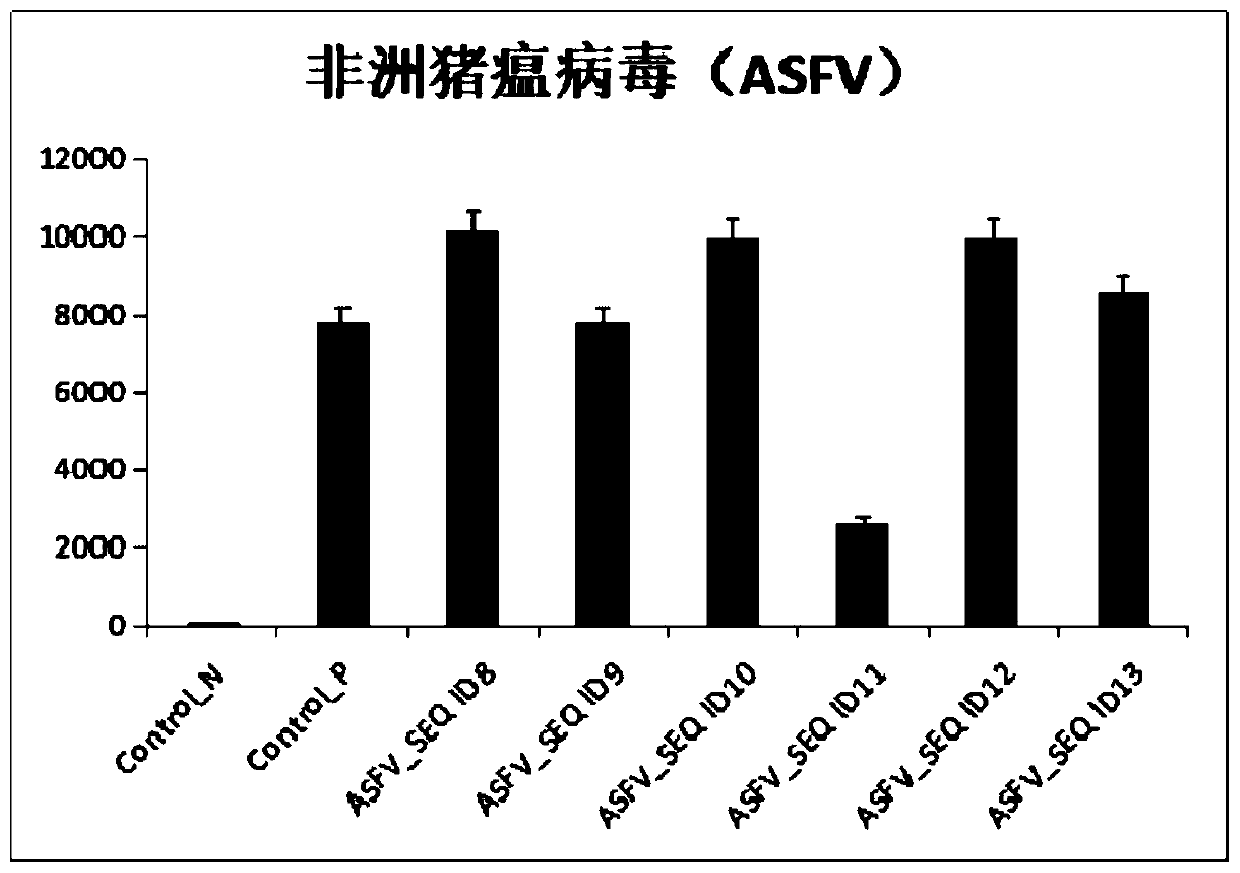
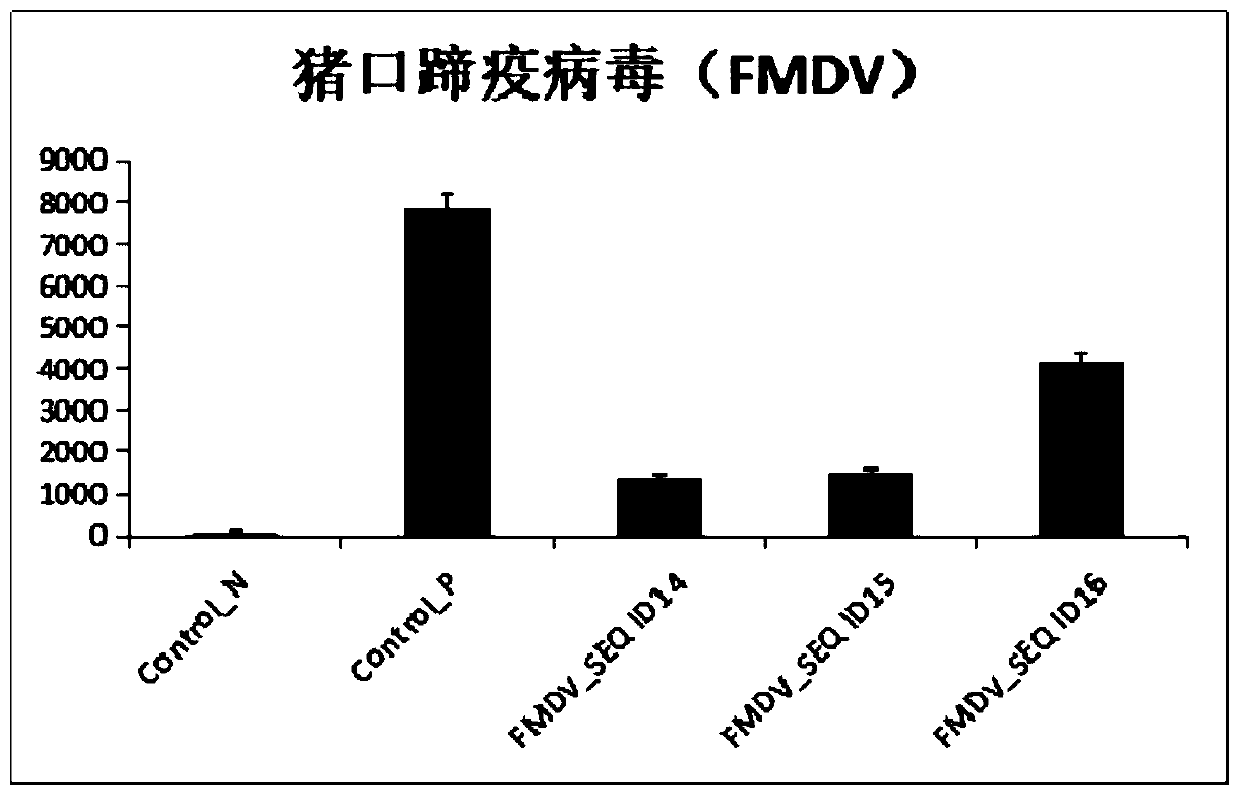
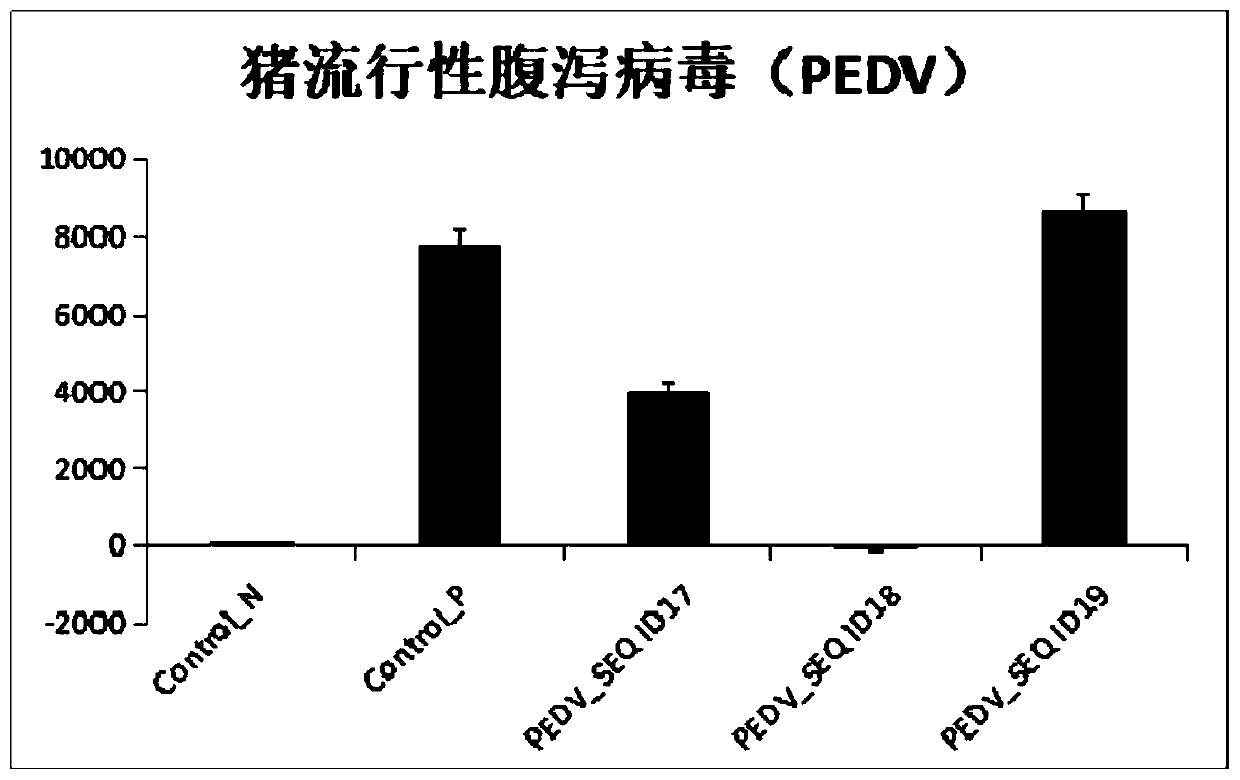
[0071] In order to realize the simultaneous detection of multiple pathogenic bacteria, we have developed a multiplex detection system for the above pathogenic targets. Since African swine fever virus is a DNA virus among the pathogens of porcine infectious diseases, the other four viruses are all RNA viruses, and CRISPR / Cas12a detection can only be performed after RT-RPA. The identified regions of the genomes of these four pathogens and the corresponding gRNA sequences were analyzed. According to parameters such as sequence similarity, GC content, base homogeneity, presence or absence of secondary hairpin structures, and presence or absence of cross-reaction in the same reaction, etc. The reaction system and gRNA combination method were optimized.
[0072] 1. Specific operation: After preparing the gRNA according to the method of step 3 in Example 2, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com