Method for improving citrus canker resistance based on CRISPRCas9 mediated CsWRKY22 site-directed editing
A citrus canker and editing technology, applied in the field of plant genetic engineering, can solve the problems of citrus canker resistance that needs further research, enhance canker susceptibility, etc., and achieve the effect of enhancing citrus canker resistance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] 1. Extraction of DNA
[0037] Select 0.1-1 g of citrus veins, use Aidlab company kit (cat. No. DN15) to extract genomic DNA, and store at -20°C for future use.
[0038] 2. PCR Amplification of Genomic Sequence
[0039]
[0040] The amplification program was: 94°C, 5min; 94°C, 30sec, 56°C, 30sec, 72°C, 1.5min, 35 cycles; 72°C extension, 10min.
[0041] 3. DNA fragment recovery, ligation, transformation into Escherichia coli DH5α
[0042] Under the ultraviolet light, use a clean blade to cut off the agarose gel block containing the fragment of interest. The gel recovery method was carried out according to the instructions of the kit (purchased from Omega Company), and the recovered fragments were quantified by electrophoresis on agarose gel.
[0043] The establishment of the enzyme digestion system and the reaction conditions were carried out with reference to the instructions of the restriction endonuclease kit from TaKaRa Company. The recovered fragments were dig...
Embodiment 2
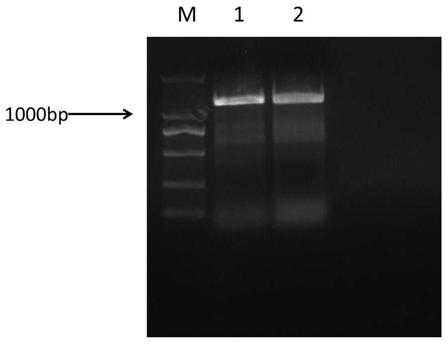
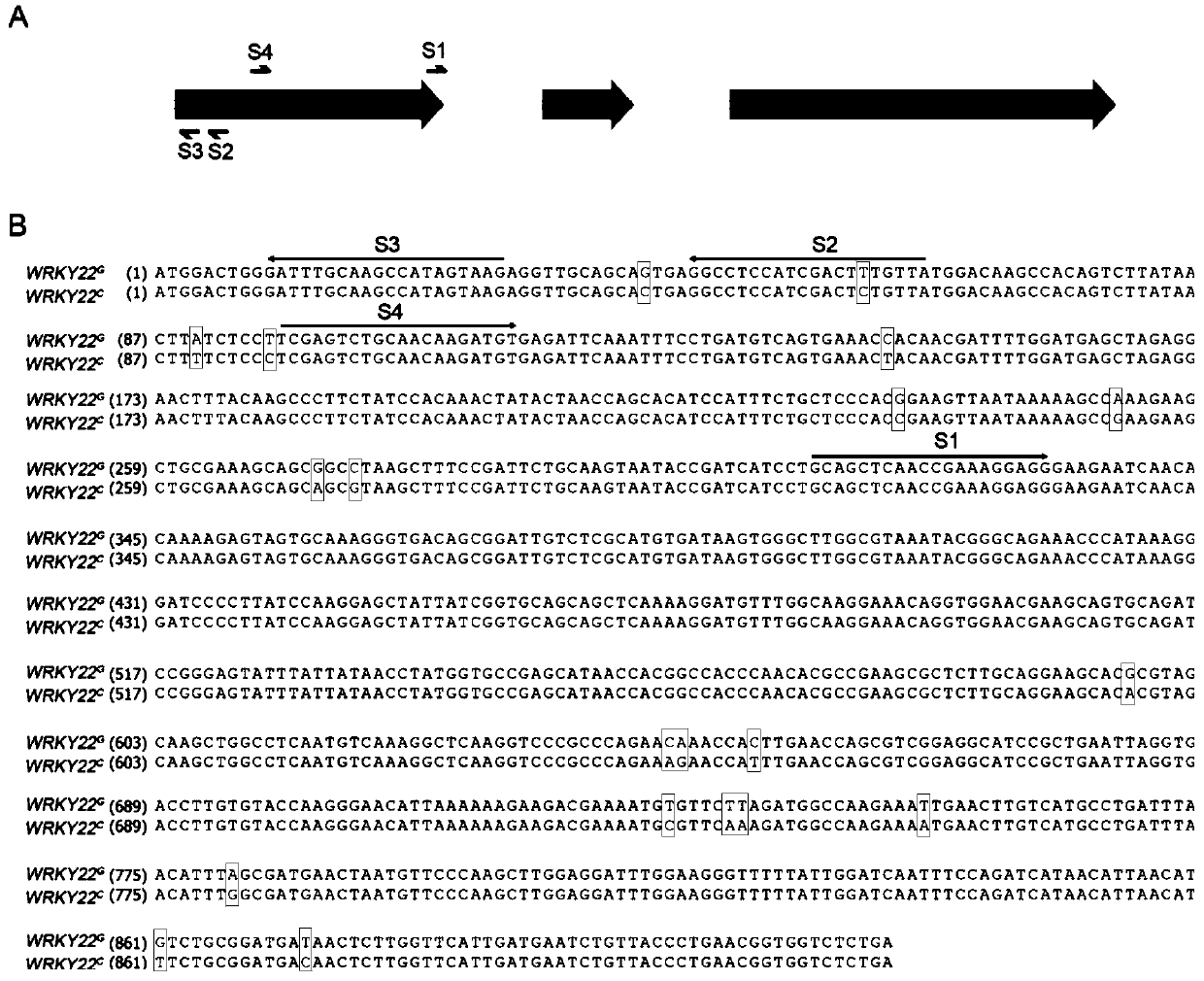
[0052] Using Wanjincheng genome as a template and WRKY22-f / WRKY22-r as primers (Table 6), the full-length gene of Wanjincheng CsWRKY22 was amplified by PCR, and the electrophoresis band of the target product was about 1.2kb ( figure 1 ), sequencing analysis found that the Wanjincheng CsWRKY22 gene contains three exons and two introns, and there are two alleles in Wanjincheng, and the coding sequence contains abundant single base polymorphism SNP, Based on the first SNP "G / C" difference, the two alleles were named CsWRKY22 respectively G and CsWRKY22 C ( figure 2 ). No insertions or deletions were detected, suggesting that both alleles encode the same functional WRKY22 protein.
[0053] Large-scale (more than 27 clones) sequencing analysis of T clones on the CDS of Wanjincheng Cs WRKY22 coding sequence, three repeated experiments showed that CsWRKY22 G and CsWRKY22 C The heterozygosity in the Wanjincheng genome is 3:1, that is, there are three copies of CsWRKY22 in the Wa...
Embodiment 3
[0055] 1. Design of sgRNA
[0056] Based on the WRKY22 genome ( figure 2 ) and protein sequence ( image 3 ), the sequences that perform the biological functions of WRKY22 are located on the second and third exons, so in order to effectively inactivate the WRKY22 protein function, the present invention uses the first exon sequence of WRKY22 as a target to screen potential Cas9 targets. Four (S1 / S2 / S3 / S4) sgRNAs were selected according to the location and GC content of the target (Table 2). The target sequences are S1: 5'-CCTCTCTTTCGGTTGAGCTGC-3'; S2: 5'-GGCCTCCATCGACTTTGTTA-3'; S3: 5'-GATTTGCAAGCCATAGTAAG-3'; S4: 5'-ACATCTTGTTGCAGACTCGA-3'. The analysis and evaluation results of the four sgRNA targets are shown in Table 2.
[0057] 2. Identification of sgRNA activity
[0058] To verify the in vitro activity of the above sgRNA, the operation steps are as follows:
[0059] (1) sgRNA reverse transcription in vitro.
[0060] The sgRNA in vitro transcription kit (Novoprotein...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com