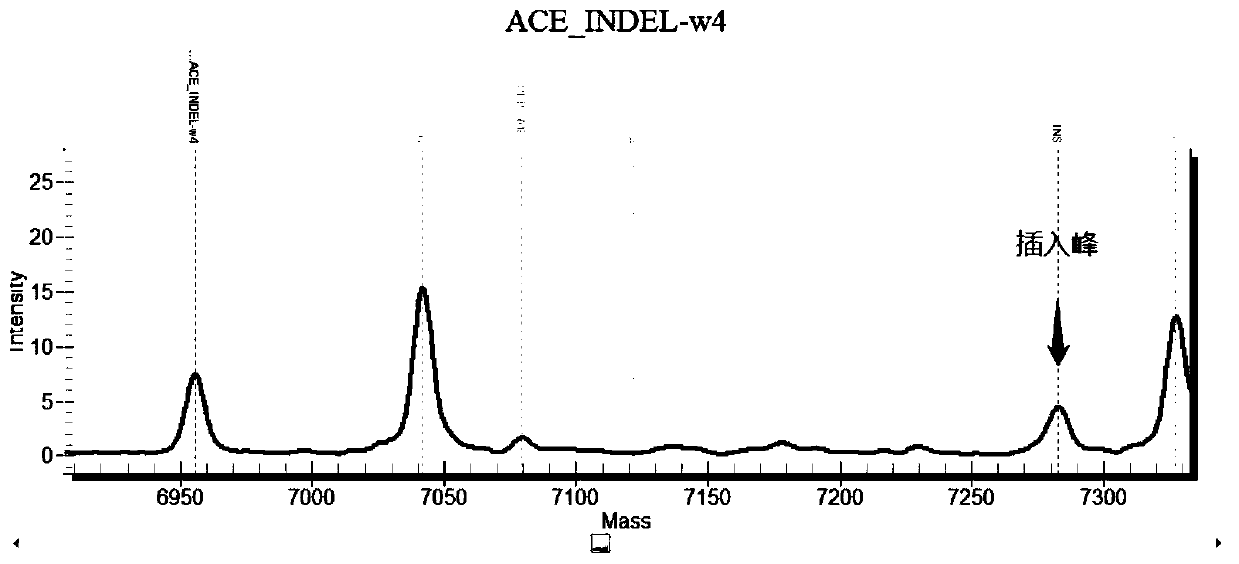
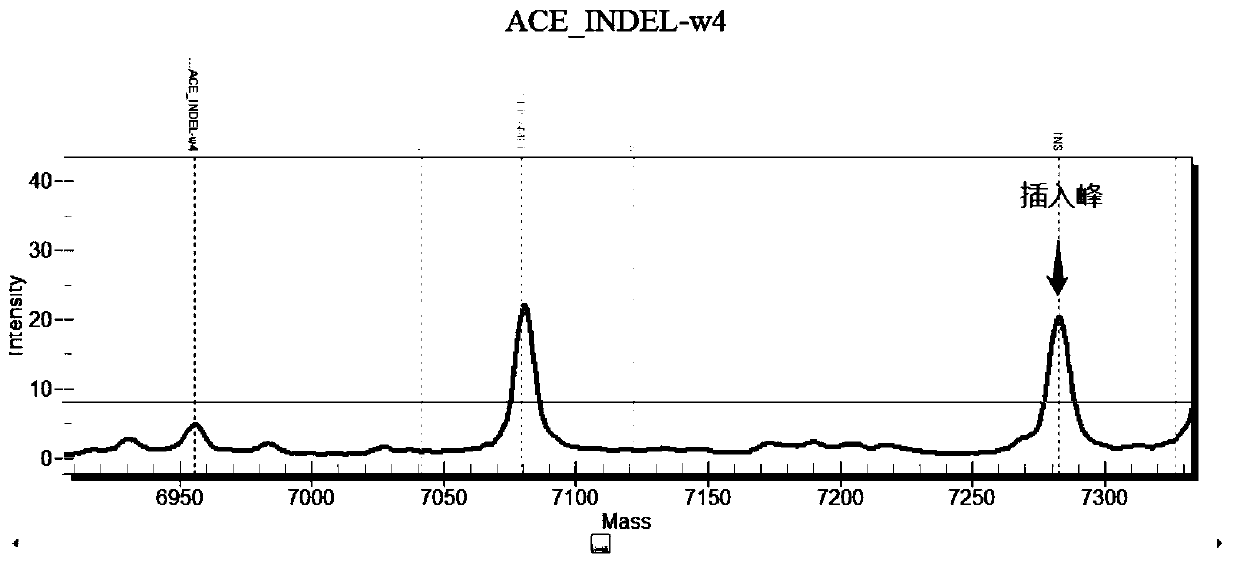
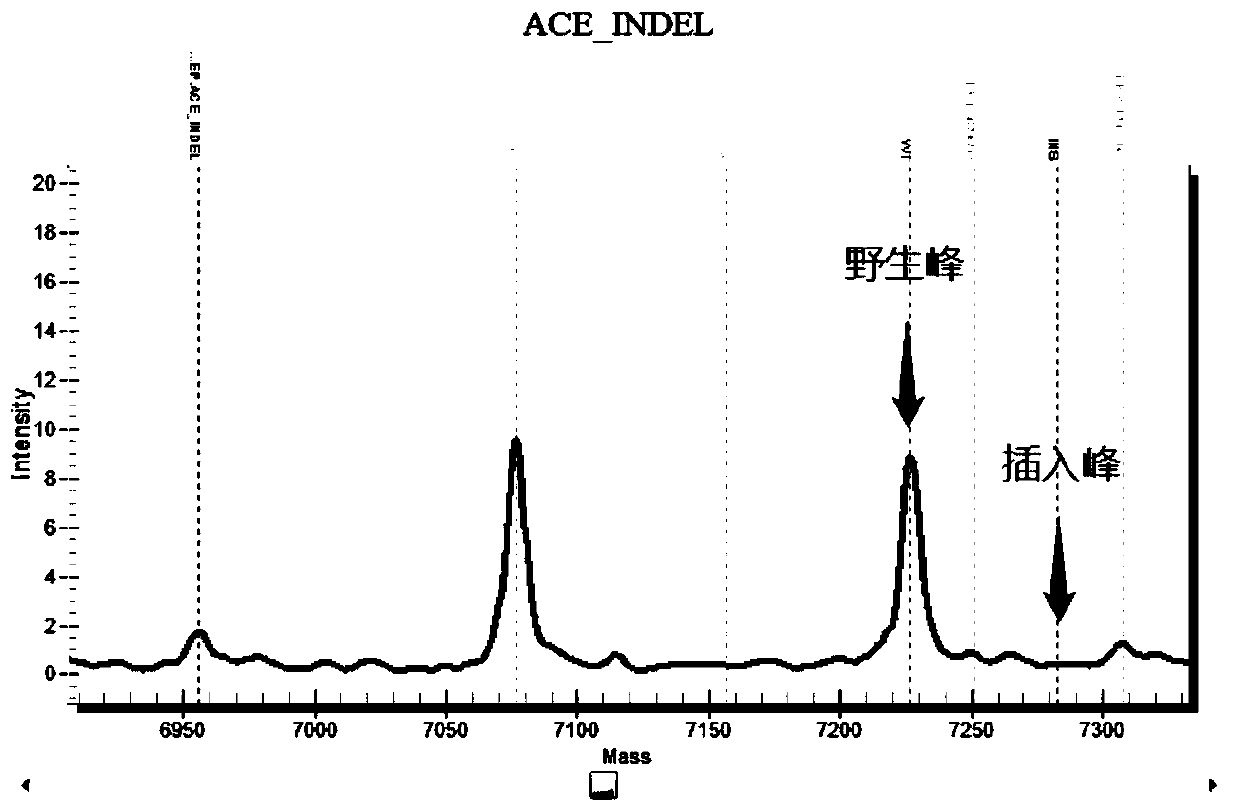
Chromosome long segment insertion detection method and long segment insert detection method based on MassARRAY platform
A detection method and long-fragment technology, applied in the biological field, can solve the problems of low cost, low throughput and accuracy, and achieve the effects of low consumption, reduced false positive results, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0130] Taking the ACE gene rs4646994 as an example, rs4646994-F is a shared forward primer, rs4646994-R1 is a wild-type reverse primer, and rs4646994-R2 is an insertion-type reverse primer; the wild-type and insertion-type single-base extension primers are the same.
[0131] Table 1 PCR primer sequences
[0132] PCR_ID PCR_SEQ rs4646994-F ACGTTGGACTGGAGACCACTCCCATCCTTT rs4646994-R1 ACGTTGATGTGGCCATCACATTCGTCAGAT rs4646994-R2 ACGTTGATTGAGACCATCCCGGCTAAAACG
[0133] Table 2 Sequences of single-base extension primers
[0134] SNP_ID UEP_SEQ rs4646994 GACCTGCTGCCTATACAGTCACTTTTT
[0135] 1. gDNA extraction: Initial peripheral blood input volume was 200 μl, extracted with QIAamp DNA Mini Kit, and eluted with 50 μl TB;
[0136] One oropharyngeal sampler, wipe the mouth 40 times, use Hi-Swab DNA Kit to extract, and 50 μl TB to elute;
[0137] 2. PCR process
[0138] (1) Dilute the sample to 10ng / μl;
[0139] (2) Prepare ...
experiment example
[0184] This experimental example mainly verifies the accuracy, specificity, sensitivity and precision of the double-hole detection method applied in the present invention.
[0185] The accuracy verification scheme of this experimental example: 20 cases were detected at each site, and compared with Sanger sequencing, the expected target was 95%.
[0186] The specific verification scheme of this experimental example: Included in the accuracy, the expected target is 95%.
[0187] The sensitivity verification scheme of this experiment example: included in the accuracy, the expected target is 95%.
[0188] The precision verification scheme of this experimental example (including intra-batch, inter-batch, and personnel comparison, not involving inter-instrument comparison) is shown in Table 6 below:
[0189] Intra-batch precision: each sample was repeated 3 times in the same batch to compare the intra-batch precision;
[0190] Batch-to-batch precision: the same operator tests the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com