Low-yield higher-alcohol beer yeast strain and construction method thereof
A technology of brewer's yeast and brewer's yeast, which is applied in the field of bioengineering, can solve the problems of large difference in regulation effect, reduction of higher alcohol content, unsatisfactory application, etc., achieve good fermentation performance and growth performance, and lower higher alcohol effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Construction of low-yield higher alcohol brewer's yeast engineering strains
[0056] The starting strain S. cerevisiae S17 was constructed by homologous recombination to construct recombinant genetically engineered strains.
[0057] According to the yeast genome data in Genebank and the integrated plasmid sequence, the primers in the following examples were designed.
[0058] The PCR primers used in the present embodiment of table 1
[0059]
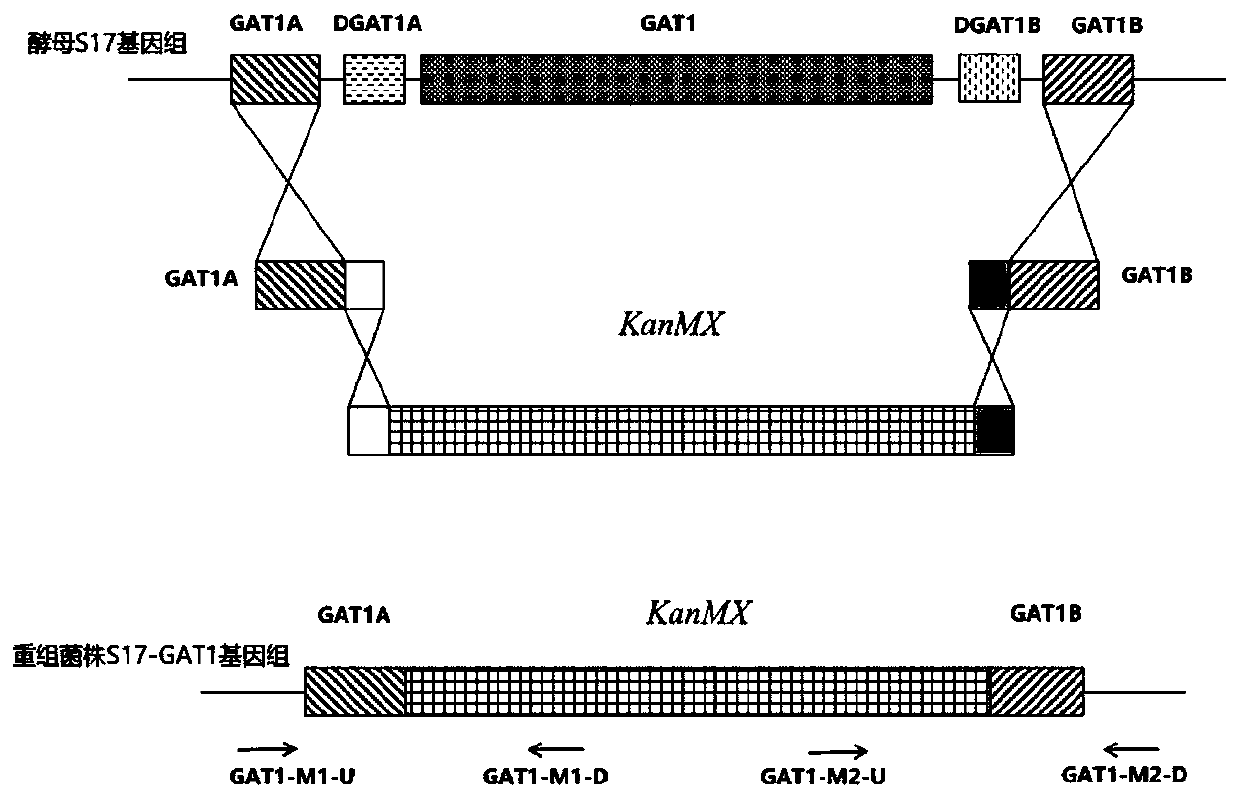
[0060] The main construction process of the strain is as follows (homologous recombination strain construction flow chart is attached figure 1 shown):
[0061] 1) Amplification of the fragment required for one allelic knockout of the GAT1 gene
[0062] Using the genome of Saccharomyces cerevisiae S17 as a template, using GAT1A-F and GAT1A-R as primers to PCR amplify an allelic knockout upstream homologous sequence GAT1A fragment of the GAT1 gene, the length is 564bp; using the genome of Saccharomyces cerevisiae S1...
Embodiment 2
[0081] Example 2: Fermentation experiment of recombinant strain S17-DΔgat1-k-p wheat beer
[0082] 1) Fermentation process roadmap: refer to the attached Figure 10 .
[0083] 2) Process conditions: crushing conditions: the degree of crushing is suitable for wheat malt without whole grains, and the degree of crushing is not too fine, so as not to cause excessive filtration pressure; liquefaction and saccharification conditions: the crushed wheat malt is mixed with a material-to-water ratio of 1:4 Proportionally add warm water at 30°C, stir well, place in a constant temperature water bath, keep at 30°C for 30 minutes, raise the temperature to 65°C at 2.0°C / min, keep for 90 minutes, rapidly raise the temperature to 78°C, and keep for 10 minutes. Fully stir once every 5 minutes during the saccharification process; filter conditions: filter the wheat wort after saccharification while it is hot, and wash the lees with hot water at 75°C for 3 times; ‰ of bitter hops (based on malt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com