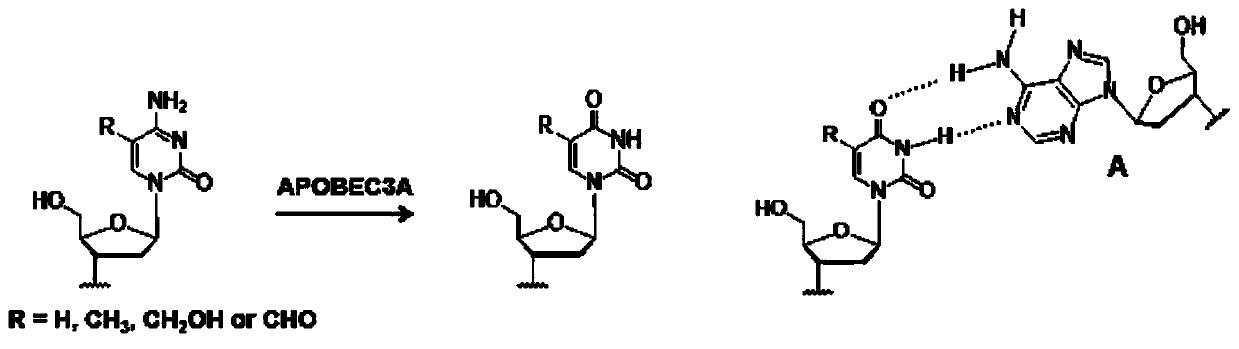
Single-base resolution localization analysis method for 5-carboxycytosine modification in DNA assisted by deaminase
A carboxylcytosine and analysis method technology, which is applied in the field of 5-carboxycytosine modified single base resolution localization analysis, can solve the problems of increasing purification steps, DNA loss and the like, achieves high deamination efficiency, is easy to operate, and is conducive to localization The effect of analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0045] Standard Synthetic DNA Analysis
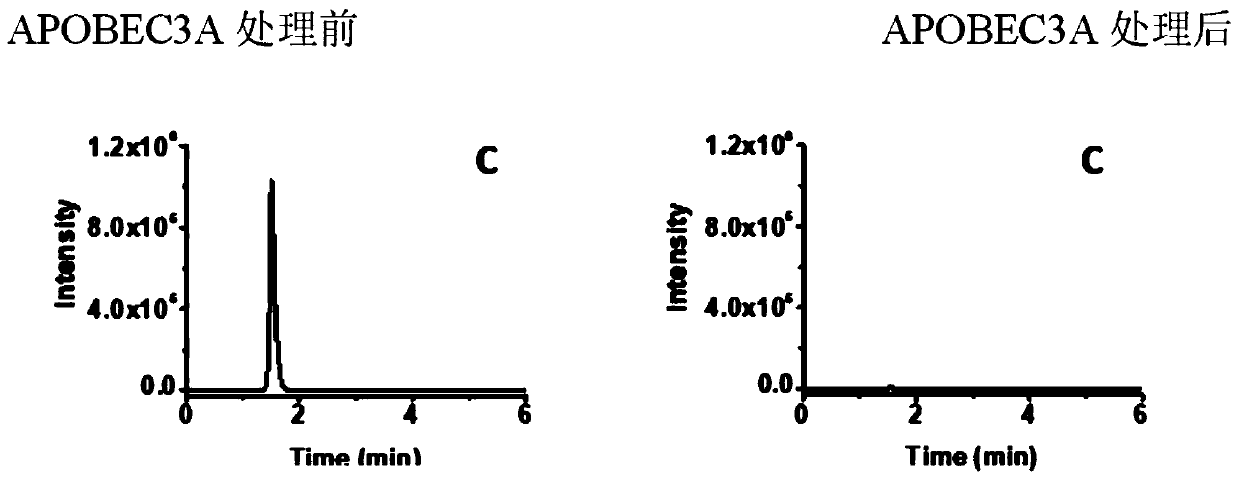
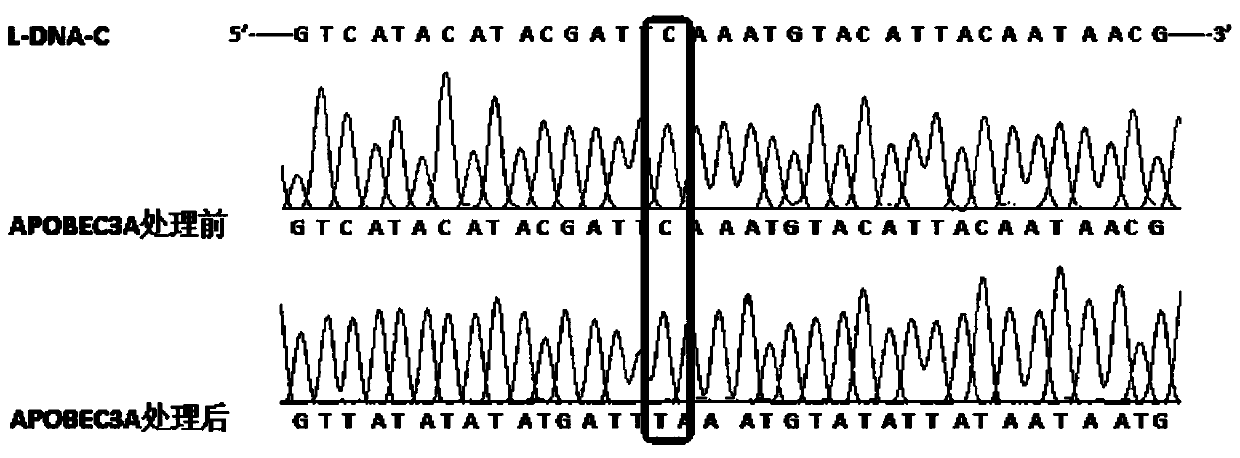
[0046] Commercially synthesized DNA 100ng containing C, 5-mC, 5-hmC, 5-fC and 5-caC, respectively, was added with a certain amount of APOBEC3A protein (according to the actual activity of the APOBEC3A protein used to determine the most suitable deamination concentration ), 2 μL of 250 mM HEPES, add deionized water to make the reaction system 20 μL, and incubate in a 37°C water bath for 2 hours. This was followed by incubation for 10 minutes in a 90°C water bath.
[0047] The above reaction DNA was taken for polymerase chain amplification reaction. Reaction system: 2 μL of 10x amplification buffer, 1 μL of 10 μmol / L forward and reverse primers, 5 ng of template DNA, 10 U of taq DNA polymerase final concentration, add deionized water to make the system 20 μL. The annealing temperature of the amplification reaction is selected according to different regions; the amplification cycle time program: (1) denaturation at 95°C for 5min; (2) den...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com