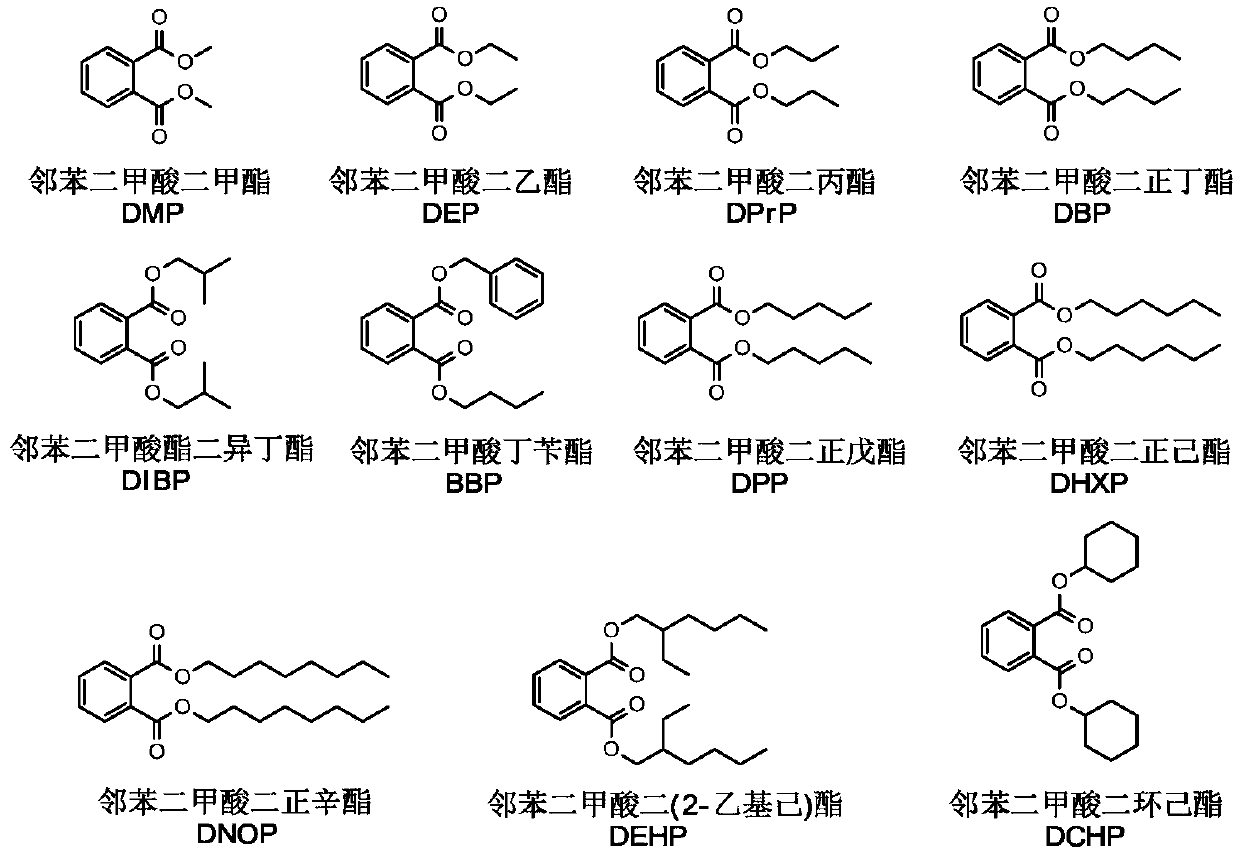
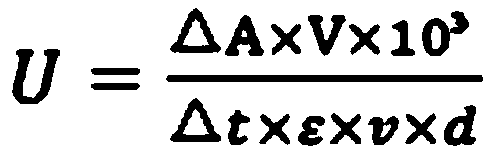DEHP (di-2-ethylhexyl phthalate) hydrolase, gene and application of hydrolase
A kind of phthalate, hydrolase technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Gene Cloning of DEHP Hydrolase GoEst15
[0047] According to the open reading frame of DEHP hydrolase GoEst15, the upstream and downstream primers were designed, and the genomic DNA of Gordonia polyisoprenivorans was used as a template for PCR amplification.
[0048] The designed upstream and downstream primers are as follows:
[0049] Upstream primer SEQ ID No.3:
[0050] 5'-G GAATTC ATGGGAGTTCCGAACACC-3';
[0051] Downstream primer SEQ ID No.4:
[0052] 5'-CCC AAGCTT TCAGACGTCGAGCGCGGC-3';
[0053] Wherein, the underlined part of the upstream primer is the cleavage site of the restriction endonuclease EcoR I, and the underlined part of the downstream primer is the cleavage site of the restriction endonuclease Hind III.
[0054] The PCR system was: 2×Taq PCR MasterMix 25 μL, 0.5 μL each of upstream primer and downstream primer (100 μM), 1 μL genomic DNA of Gordonia polyisoprenivorans (100 ng / μL), and 23 μL wxya 2 O. The PCR amplification program is:...
Embodiment 2
[0055] Example 2 Preparation of DEHP hydrolase GoEst15 recombinant expression plasmid and recombinant expression transformant
[0056] The target DNA fragment of DEHP hydrolase obtained by PCR amplification in Example 1 and the pET28a empty plasmid were double-digested with restriction endonucleases EcoR I and Hind III at the same time, then purified by agarose gel electrophoresis, and recovered by a DNA kit. The recovered enzyme-digested target fragment and empty vector were separated at T 4 Under the action of DNA ligase, they were ligated at 4°C for 16 hours to obtain the recombinant plasmid pET28a-GoEst15.
[0057] Transform the resulting recombinant plasmid into E.coli DH5α, spread it on an LB medium plate containing 50 μg / mL kanamycin, and incubate at 37°C for 12 hours. Perform colony PCR verification on the grown colonies, and pick and amplify successfully A positive clone with a target band of about 1620bp in length was obtained. After sequencing and verification, the ...
Embodiment 3
[0058] Example 3 Induced expression of DEHP hydrolase GoEst15
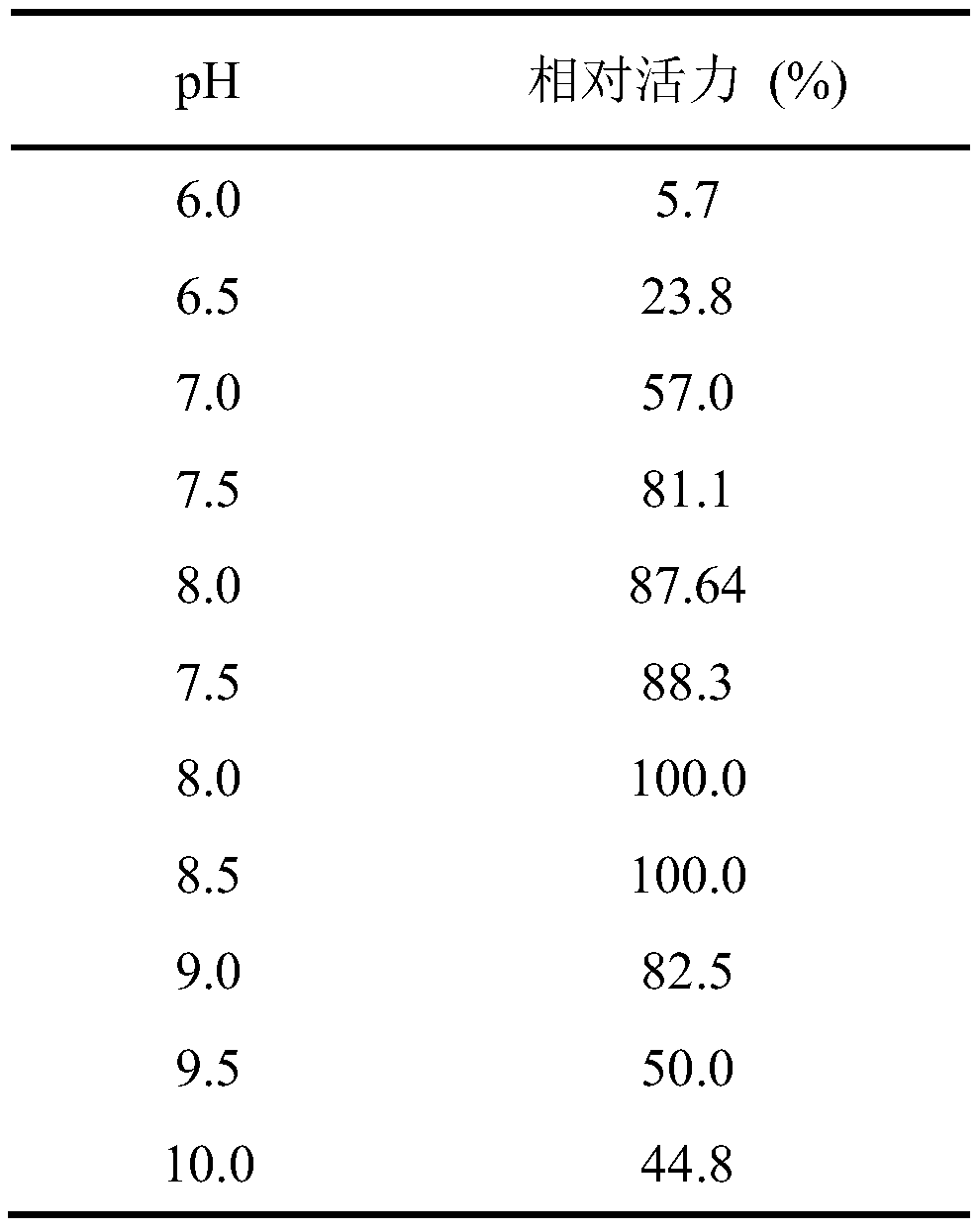
[0059] The recombinant expression transformant E.coli BL21(DE3) / pET28a-GoEst15 obtained in Example 2 was inoculated into LB medium containing 50 μg / mL kanamycin, shaken at 37°C for 12 hours, and then pressed Inoculate 1% (v / v) of the inoculum into a 250mL Erlenmeyer flask containing 50mL of LB medium (containing 50μg / mL kanamycin), put it in a shaker, and culture it with shaking at 37°C and 200rpm. OD of liquid 600 When the temperature reached 0.6, IPTG was added to a final concentration of 1 mmol / L for induction. After induction at 16°C for 24 hours, the culture medium was centrifuged at 12,000 rpm to collect cell pellets and washed with saline to obtain resting cells. Suspend the resting cells obtained by the above method in 10 mL of potassium phosphate buffer (100 mM, pH 7.0), ultrasonically break in an ice-water bath, and centrifuge to collect the supernatant, which is the crude enzyme solution of recombinant...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com