Novel method for protein O-GalNAc modification rapid library search and deep coverage
A protein and mass spectrometry technology, applied in the field of bioinformatics, can solve problems such as large search space, missing secondary spectra, and limited enrichment strategies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, urinary protein O-GalNAc spectrum classification search
[0054] Step 1. Urinary protein extraction and enzyme digestion
[0055]Take 10mL of the middle morning urine of healthy people, centrifuge at 12000g for 15 minutes to remove impurities, take the supernatant, transfer it to a 50mL centrifuge tube, add 3 times the volume of pre-cooled acetone, mix well, and then stand at -20°C for 2-4 hours for urine analysis. protein precipitation. After the precipitation is completed, centrifuge at 12000g for 15 minutes, collect the precipitated part, add lysate (8M urea, 0.1M Tris-HCl, pH=8.5) after drying, and use an ultrasonic breaker to sonicate urine protein (30% power, each time Ultrasound for 2 seconds, repeated 10 times), after the end of the ultrasound, centrifuge at 16000g for 15 minutes, and take the supernatant, namely the urine protein extract. Dithiothreitol was first added to the obtained urine protein extract to make the concentration 10mM (the fun...
Embodiment 2
[0064] Example 2. O-GalNAc Glycopeptide Matching and Quantitative Missing Value Filling Using Chromatographic Retention Time Correction and Accurate Charge-to-Mass Ratio
[0065] Step 1-step 4 is the same as embodiment 1.
[0066] Step 5. Chromatographic retention time correction and quantitative missing value filling
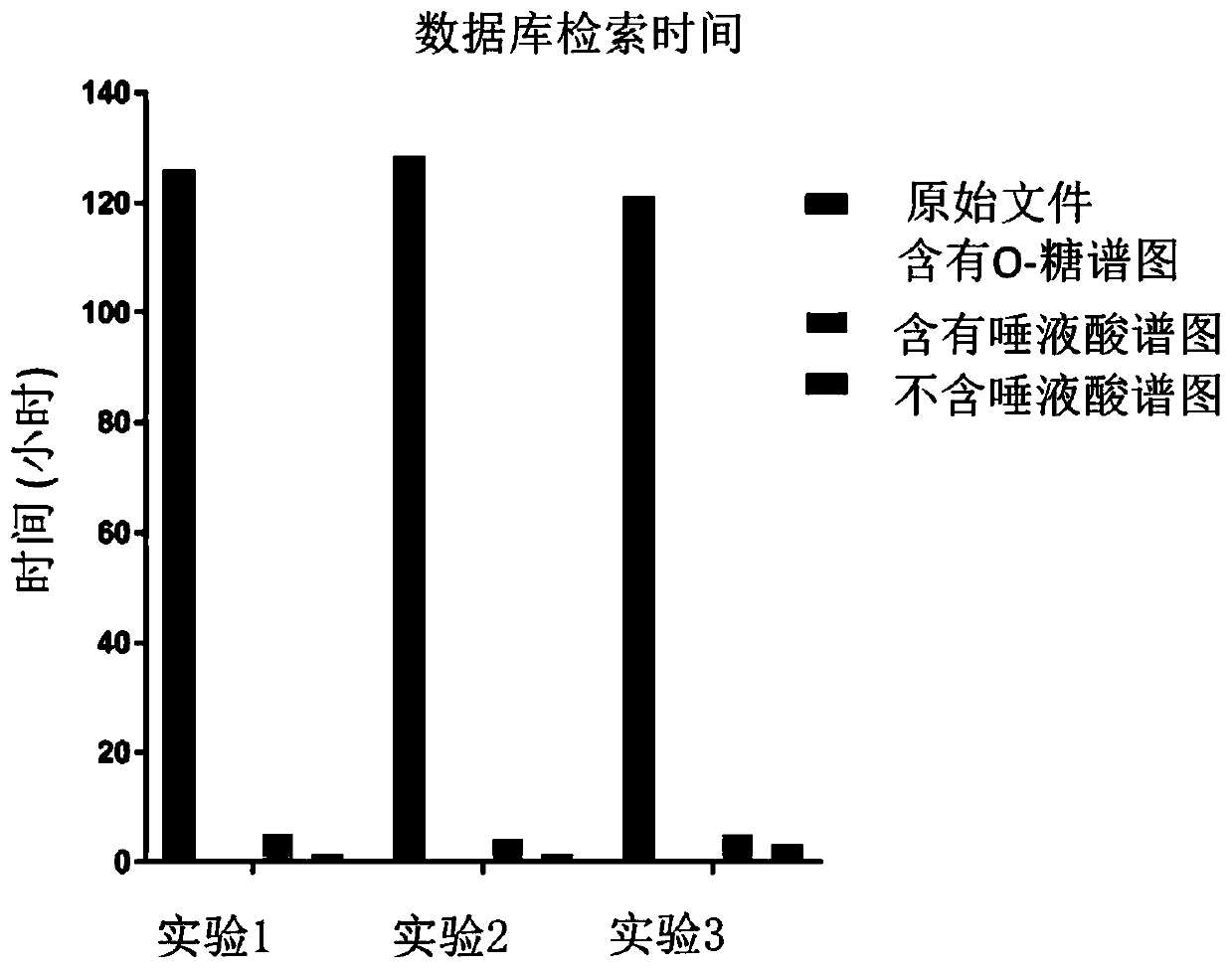
[0067] After searching the Byonic database and extracting information from the raw files of 16 healthy male samples and 20 healthy female samples, the retention time prediction of the identification results in 36 samples was carried out. The correction method is as follows: First, the O-GalNAc glycopeptides identified at least 3 times in 36 files (identified in at least 3 samples out of 36 samples) were identified as high-confidence peptides, and quantitative analysis was performed on them. A total of 487 high-confidence peptides were found, and the reference retention times of these 487 O-GalNAc glycopeptides were calculated (see above for specific methods). ...
Embodiment 3
[0069] Example 3. Using chromatographic retention time correction and accurate charge-to-mass ratio for O-GalNAc glycopeptide matching and quantitative missing value filling accuracy verification
[0070] Step 1-step 4 is the same as embodiment 1.
[0071] Step 5. Chromatographic retention time correction and quantitative missing value filling
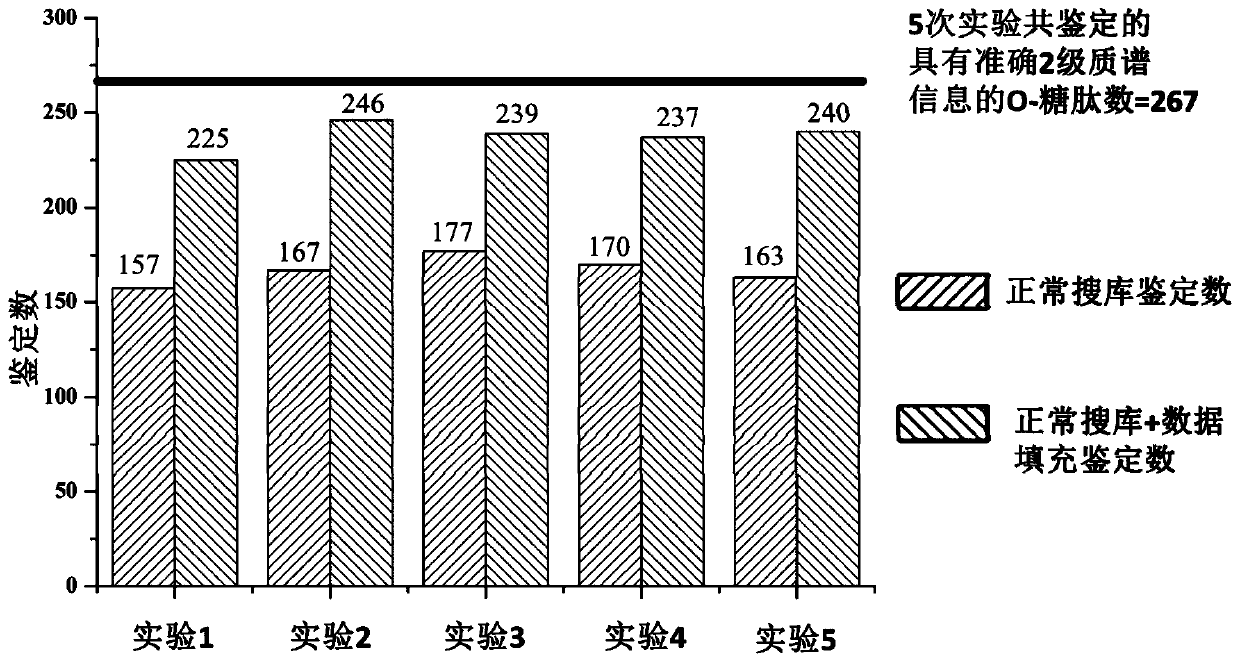
[0072]The samples from the same case of urinary protein glycopeptides were divided into 5 equal parts, and then the mass spectrometry data were collected according to the method of step 4 of Example 1. Retention time prediction of identifications in samples. The correction method is as follows: First, the O-GalNAc glycopeptides with MS / MS information identified in the 5 files are identified as high-confidence glycopeptides, and quantitative analysis is performed on them. A total of 267 high-confidence glycopeptides were found, and the reference retention times of these 267 O-GalNAc glycopeptides were calculated respectively (see abov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com