Metagenome or macrotranscriptome sequencing data automatic analysis method and system
An automated analysis and macro-transcriptome technology, applied in the field of automatic analysis of metagenomic or macro-transcriptome sequencing data, can solve problems such as slow interpretation speed affecting the ultimate delivery of pathogen detection, and it is difficult to quickly go back to historical samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] A method for automatic analysis of metagenomic or metatranscriptome sequencing data, which operates in a linux environment, and operates according to the following steps at the computer level:
[0078] (1) Prepare the required programs: auto_report.pl (for automatic interpretation), auto_report.R (for generating automatic interpretation reports).
[0079] (2) Prepare the required databases: CRP database, CCRP database, background database corresponding to each sample type, detection process, and strain type, etc.
[0080] Specifically, the CRP database (database of clinically reportable pathogens) is established by the following methods: according to the research progress or case reports of suspected pathogens in the literature, to confirm the pathogenic bacteria to establish a database of clinically reportable pathogens; for example, it can be found in NCBI Investigate the published literature in NCBI pubmed in the database, query the research progress or case reports ...
Embodiment 2
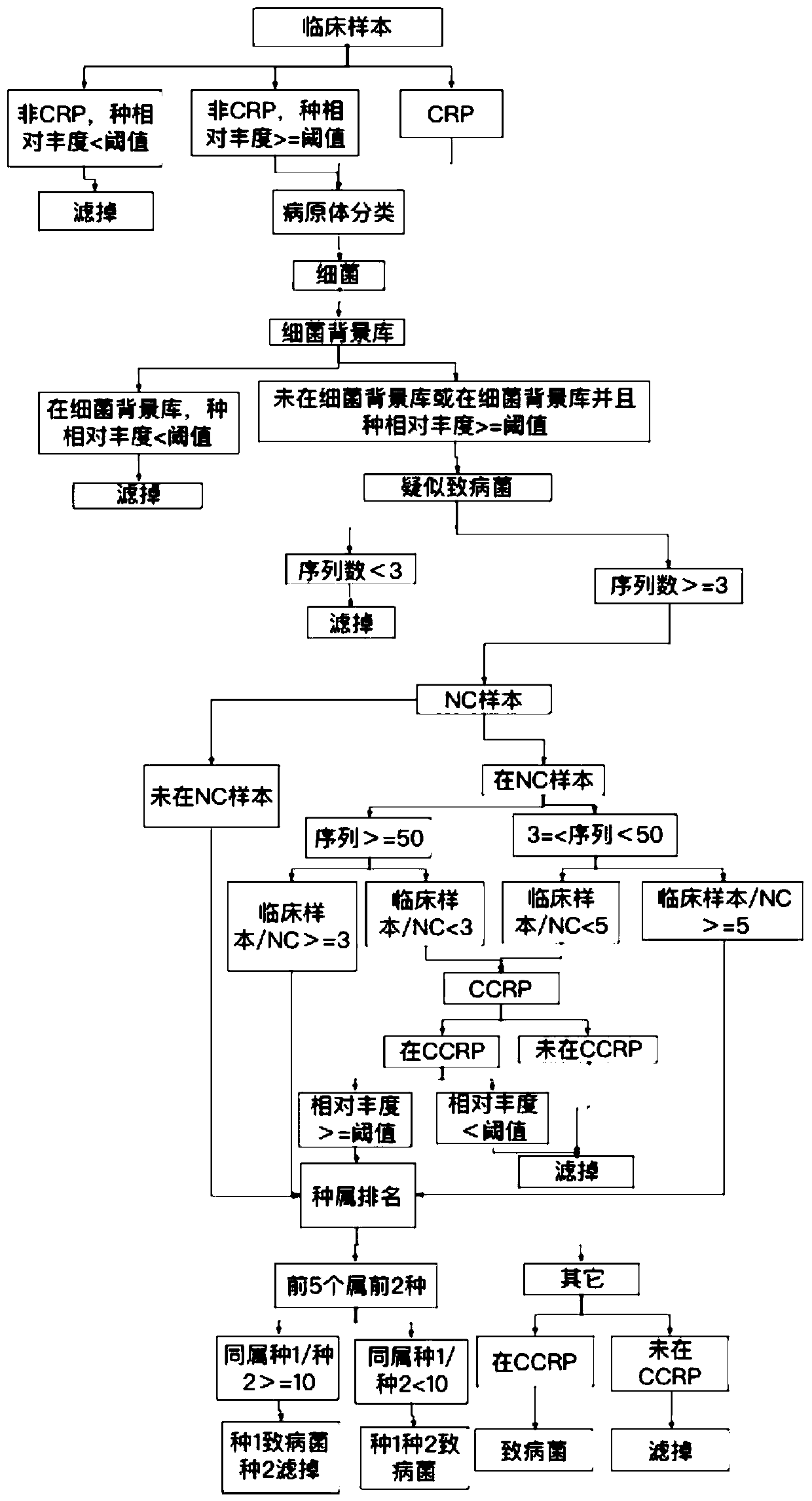
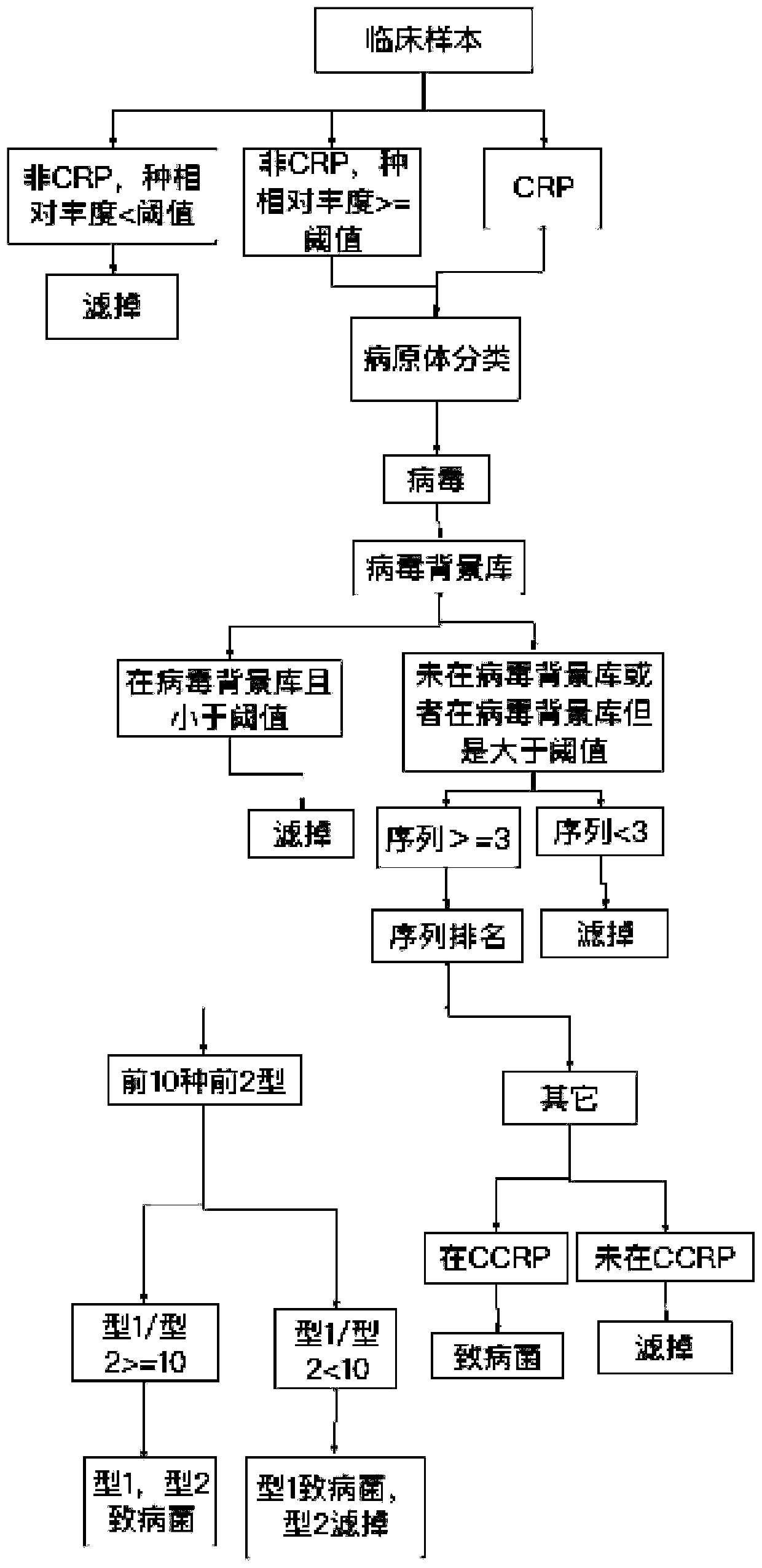
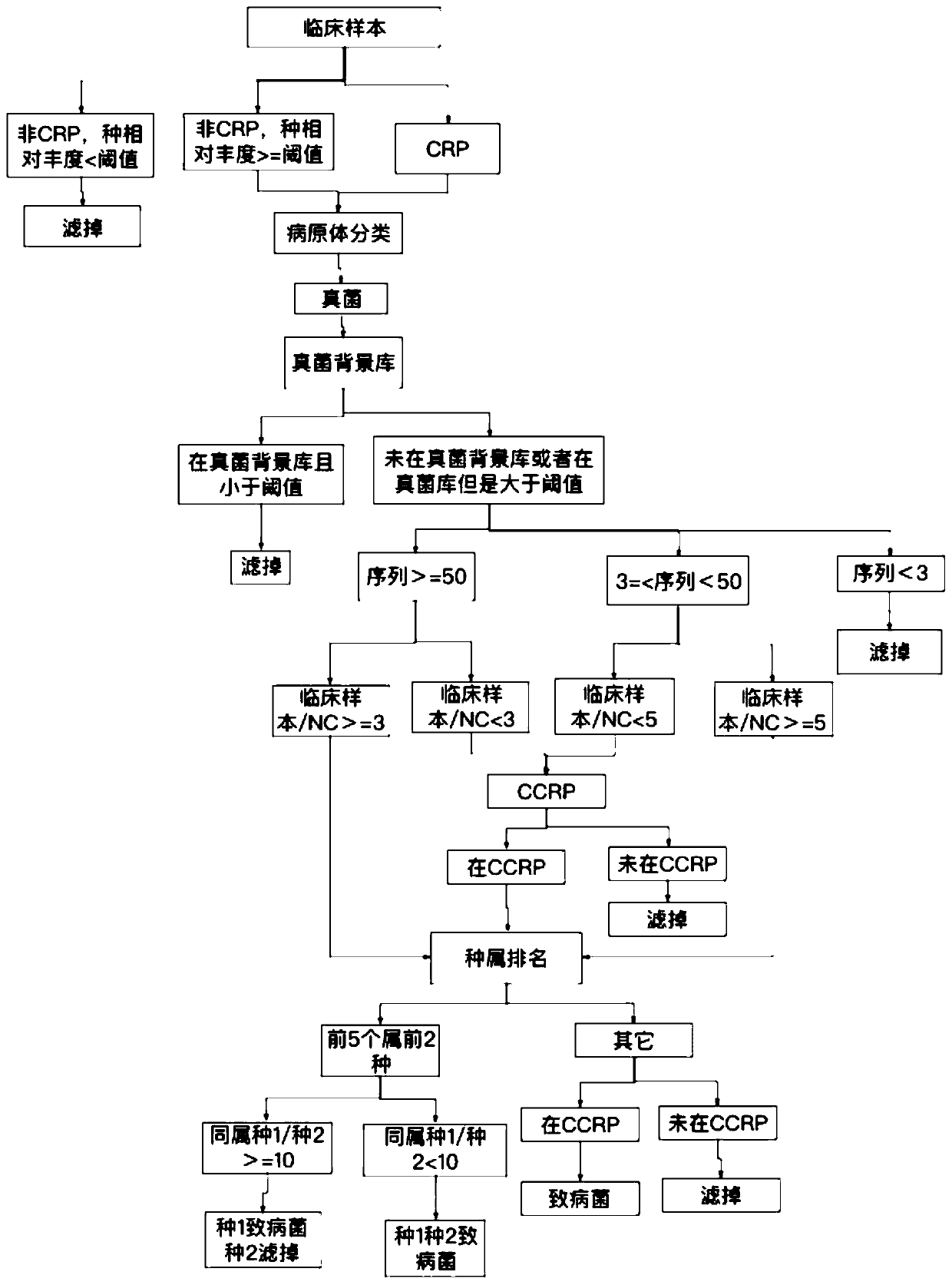
[0090] A method for automatic analysis of metagenomic or metatranscriptome sequencing data, the logic flow of which is as follows Figure 1-4 shown, including the following steps:
[0091] 1. Obtain sequencing data.
[0092] Obtain the off-machine data of the metagenomic or metatranscriptome sequencing of the sample to be analyzed, match the microbial type with the gene sequence information, and obtain the initial bacterial species list.
[0093] For example, obtain a clinical sample to be analyzed ("test1", alveolar lavage fluid), match the microbial type with the gene sequence information, and obtain the result annotation file of the information analysis: bacteria result: "test1.bac.anno", virus result: " test1.virus.anno", fungi result: "test1.fungi.anno", parasite result: "test1.parasite.anno".
[0094] According to different background library types, select the corresponding background library for comparison. Specifically, such as:
[0095] Among them, 57 results were...
Embodiment 3
[0165] The automatic analysis system is established according to the method of the above-mentioned Examples 1-3. Before the automatic analysis system is officially put into use, a systematic evaluation is required to evaluate the degree of consistency between the automatic interpretation and the manual interpretation, and whether the requirements are met.
[0166] Randomly selected 890 historical samples from 6 batches of clinical samples in our company between August 4, 2019 and August 26, 2019, which were manually interpreted, and re-interpreted using the automated analysis system of the present invention. Then, at the result level, the consistency between manual interpretation and automatic interpretation was counted, and the results are as follows:
[0167] Table 7. Consistency evaluation
[0168] consistency Number of cases percentage unanimous 881 98.98% inconsistent 9 1.02%
[0169] The consistency with manual interpretation is as high a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com