Methyltransferase and application thereof
A methyltransferase and methyltransferase technology, applied in methyltransferase and its application field, can solve the problems of flavonoid oxygen-methyltransferase not being cloned and identified, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
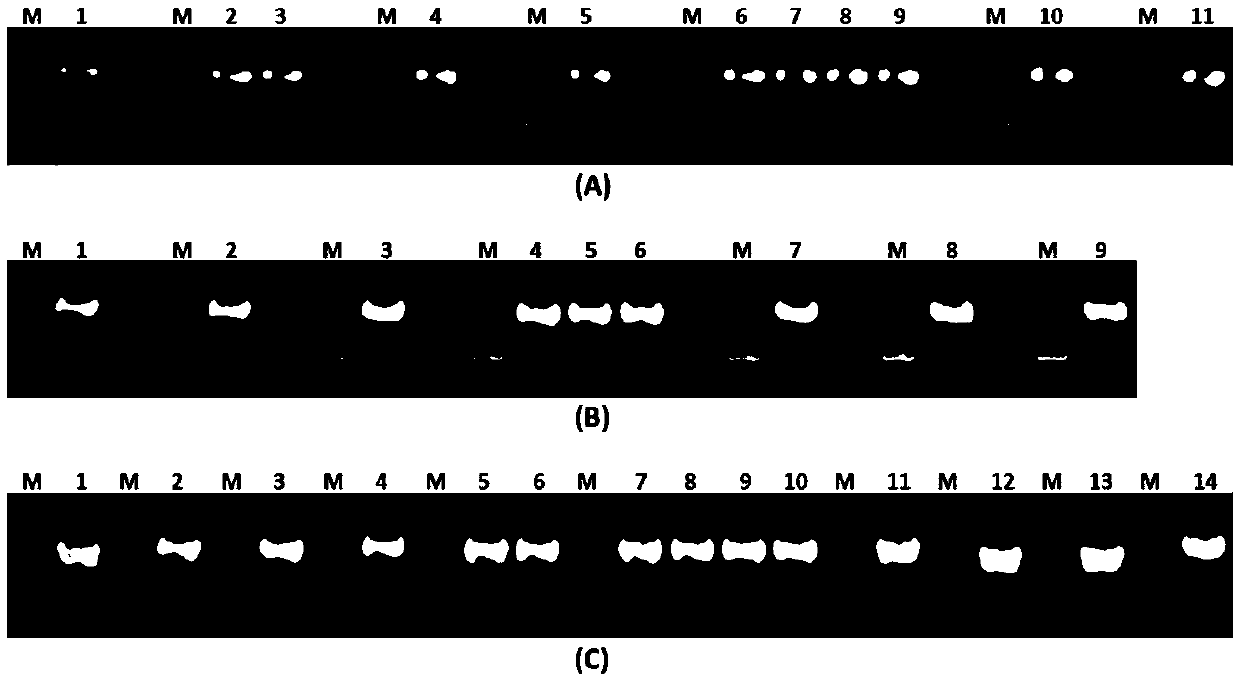
[0173] Embodiment 1, the isolation of methyltransferase and its coding gene
[0174] The inventors cloned 31 full-length cDNA sequences from soybean.
[0175] Soybean RNA was extracted and reverse-transcribed to obtain soybean cDNA. Carry out PCR amplification with this cDNA as template, use wherein primer pair 1 (SEQ ID NO:3,4); Primer pair 2 (SEQ ID NO:9,10); Primer pair 3 (SEQ ID NO:13,14) Primer pair 4 (SEQ ID NO:17,18); Primer pair 5 (SEQ ID NO:27,28); Primer pair 6 (SEQ ID NO:31,32); Primer pair 7 (SEQ ID NO:35, 36); Primer pair 8 (SEQ ID NO:39,40); Primer pair 9 (SEQ ID NO:43,44); Primer pair 10 (SEQ ID NO:51,52); Primer pair 11 (SEQ ID NO: 55,56); primer pair 12 (SEQ ID NO:59,60); primer pair 13 (SEQ ID NO:63,64); primer pair 15 (SEQ ID NO:71,72); primer pair 16 (SEQ ID NO: NO:75,76); Primer pair 17 (SEQ ID NO:81,82); Primer pair 18 (SEQ ID NO:91,92); Primer pair 19 (SEQ ID NO:95,96); Primer pair 20 ( SEQ ID NO:99, 100); primer pair 21 (SEQ ID NO: 103, 104); primer...
Embodiment 2
[0229] Embodiment 2, the construction of the Escherichia coli recombinant expression vector of methyltransferase gene GMOMT3
[0230] The plasmid GMOMT3-pMD18T containing the GMOMT3 gene constructed in Example 1 was used as a template to amplify the target gene.
[0231] The forward primer used by GMOMT3 is the 1-20 nucleotide sequence of the GMOMT3 gene, and the pGEX4T-1 homology arm sequence is added to the 5' end: GATCTGGTTCCGCGTGGATCC; the reverse primer used by GMOMT3 is the last 20 nucleotide sequence of the GMOMT3 gene , and the pGEX4T-1 homology arm sequence: GTCACGATGCGGCCGCTCGAG was added to the 5' end.
[0232] The above primers and templates were used to amplify the GMOMT3 gene by PCR method. The DNA polymerase was the high-fidelity Primer Star DNA polymerase from Treasure Bioengineering Co., Ltd., and the PCR program was set by referring to its manual: 98°C for 2min; 98°C for 15s, 58°C for 15s, 72°C for 1.2min, a total of 35 cycles; 72°C 5min at ℃; keep warm at ...
Embodiment 3
[0233] Embodiment 3, the expression of methyltransferase gene GMOMT3 in Escherichia coli
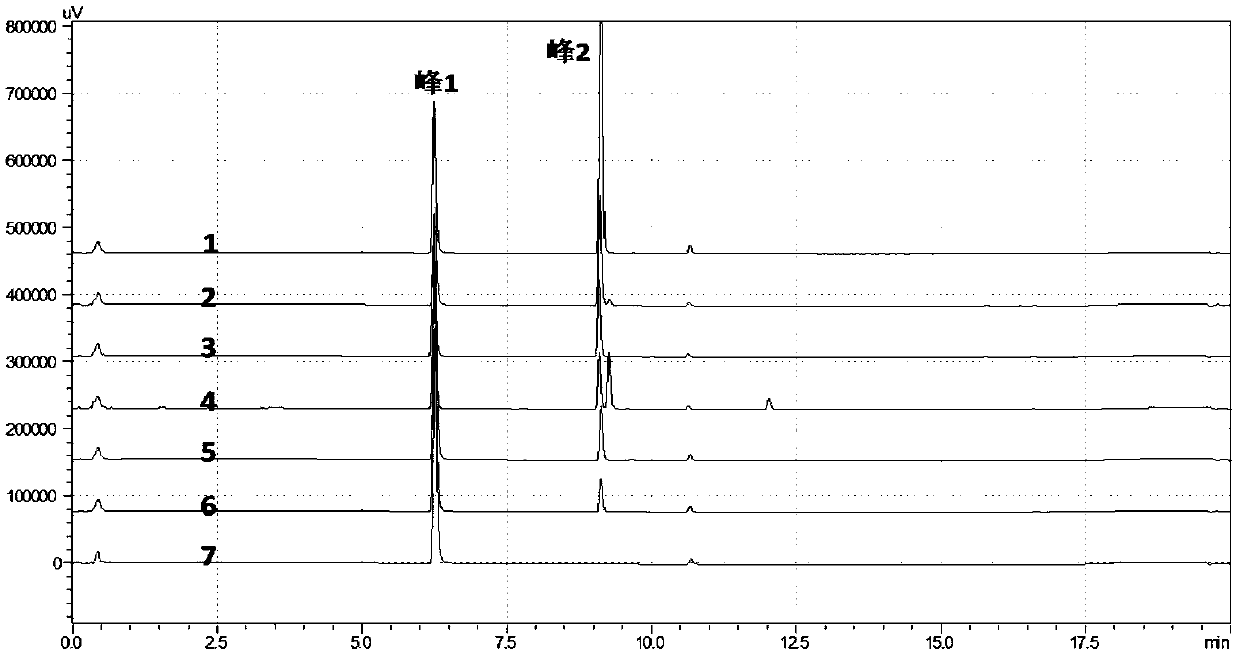
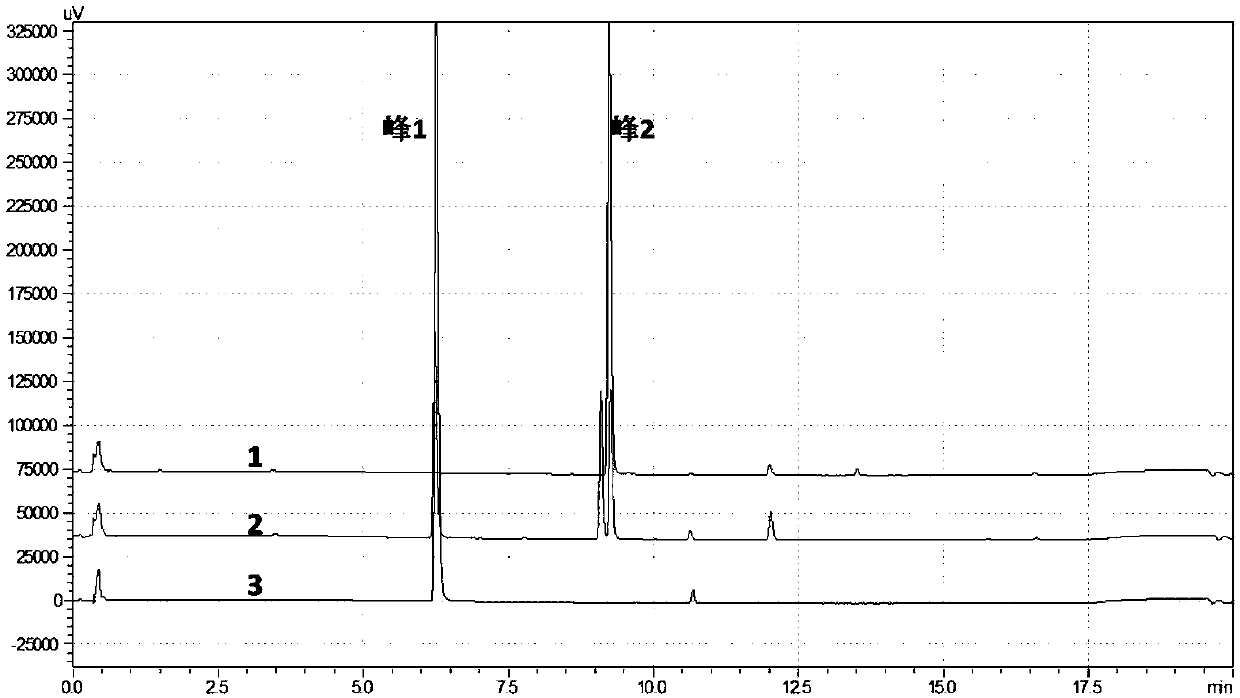
[0234] The Escherichia coli expression vector GMOMT3-pGEX4T-1 constructed in Example 2 was transformed into commercially available E.coliBL21. Inoculate a recombinant into LB medium, cultivate to OD at 37°C 200rpm 600 About 0.6-0.8, the bacterial liquid was cooled to 4°C, and IPTG with a final concentration of 200 μM was added, and the expression was induced at 18°C at 110 rpm for 18 hours. The bacteria were collected by centrifugation at 4°C, the cells were lysed with 50mM Tris-HCl, pH8.0 buffer, the supernatant of the cell lysate was collected by centrifugation at 12000g at 4°C, and samples were taken for SDS-PAGE electrophoresis. Compared with the pGEX4T-1 empty vector recombinant, the GMOMT3-pGEX4T-1 recombinant had a distinct band (approximately 65KD) characterizing GST-GMOMT3. From the results of Western Blot, it also proved that the target protein GST-GMOMT3 was soluble expres...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com