Primers for identifying types of rice GS5 gene and GLW7 gene and application of primers for identifying types of rice GS5 gene and GLW7 gene
A genotype and gene technology, applied in the field of primers to identify rice GS5 gene and GLW7 gene type, can solve the problems of high toxicity of gel components, cumbersome gel making process, and large fragment differences, so as to speed up the efficiency, process, and amplification Good effect, effect of reducing difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
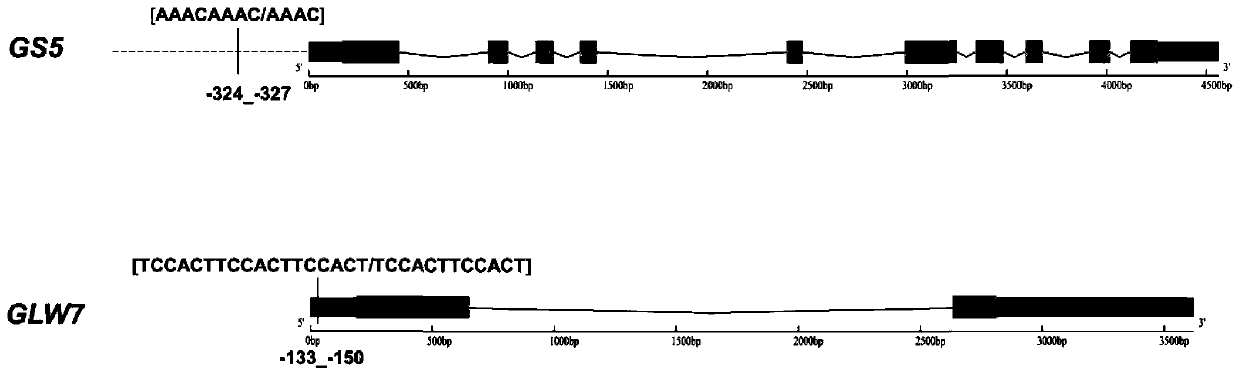
[0035] Grain shape is an important trait that determines the yield and quality of rice. Larger grain shape can increase grain weight, thereby increasing yield, while longer grain shape can improve grain filling and filling degree, reduce chalkiness, and improve rice appearance quality. GS5 and GLW7 are two key grain shape genes that have been cloned in rice, corresponding to gene numbers Os05g0158500 and Os07g0505200, respectively. The core variation is mainly reflected in the difference of the simple repeat sequence (SSR) in the promoter region or 5UTR region, such as figure 1 As shown, the functional variation of GS5 is the AAAC sequence repeat difference at 324 to 327 bp upstream of ATG, variant 1 is CAAA, corresponding to wide-grained and heavy-grained phenotypes, and variant 2 is AAACAAAC, corresponding to normal phenotypes. The functional variation of GLW7 is the TCCACT sequence repeat difference at 133 to 150 bp upstream of ATG. Variant 1 is TCCACTTCCACT, corresponding t...
Embodiment 2
[0043] In order to verify the PCR amplification and enzyme digestion identification effects of the three pairs of markers, we selected eight varieties for analysis, namely YYP1, Nipponbare (NIP), Zhenshan 97 (ZS97), Daohuaxiang (DHX), Kasalath (Kasa) , WY3, Bao Da Li (BDL) and Katy.
[0044] Accelerate the germination and germination of the above-mentioned varieties, take samples of the leaves two weeks later, put them into a 2ml centrifuge tube with steel beads, and extract DNA by TPS small-scale extraction method. The specific steps are:
[0045] 1. Shake and crush the leaves with a ball mill, add 500ul TPS buffer, and place at 65°C for 45 minutes;
[0046] 2. Centrifuge at 12000 rpm for 10 minutes, suck 300ul supernatant into a new centrifuge tube, add an equal volume of isopropanol, and place at room temperature for 45 minutes;
[0047] 3. Centrifuge at 12,000 rpm for 10 minutes to obtain a DNA precipitate, discard the supernatant, and add 500ul of 75% ethanol;
[0048] ...
Embodiment 3
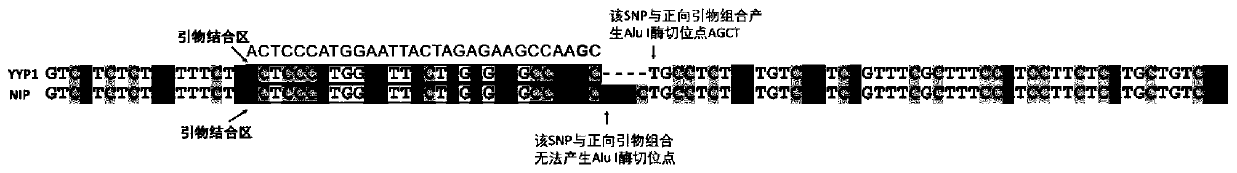
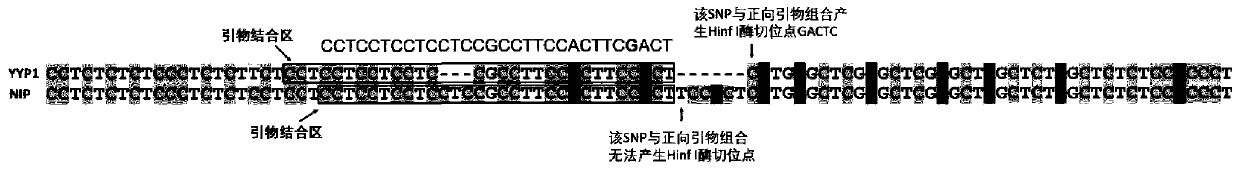
[0052] Some varieties may have variations in the primer binding sites, which may affect the amplification effect. We further clarified the identification effect of the two gene recognition primers on a large number of varieties. We collected 280 varieties distributed all over the world, planted them in the field, and extracted DNA from the leaves at the seedling stage. The extraction method is the same as above. Subsequently, the GS5 and GLW7 enzyme digestion identification markers were used for PCR amplification and enzyme digestion identification respectively, and finally the variation types of different varieties were obtained. The results of GS5 marker identification (Table 1) showed that bands were amplified in all 280 varieties, of which 52 varieties contained "AAAC" deletion wide-grain and heavy-grain variation, 224 varieties were wild type, and 3 varieties were heterozygous type, proving that the marker can be used for genotype identification of hybrid rice and F2 segr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com