Cell strain for protein display and expression as well as preparation method and application thereof
A protein display and cell technology, which is applied in the field of protein display cell lines and its preparation, can solve the problems of affecting RNA polymerase II transcription activity, complex chromosome structure, and difficulties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment one, material and method
[0030] 1. Cell culture
[0031] Both CHO-K1 cells and a series of cells constructed using CHO-K1 use PRMI-1640 medium containing 10% fetal bovine serum (Hyclone), 100U / mL double antibody, at 37°C, 5% CO 2 cultured in an incubator. CHO / dhFr- (dihydrofolate reductase-deficient Chinese hamster ovary cells) used IMDM medium containing 10% fetal bovine serum (Hyclone), 100U / mL double antibody, 0.1mM hypoxanthine and 0.016mM thymine, in 37°C, 5% CO 2 cultured in an incubator.
[0032] Human 293F cells using FreeStyle TM 293Expression Medium at 37°C, 5% CO 2 Incubator, shake flask culture at 120rpm rotation speed.
[0033] 2. Cell transfection and establishment of cell lines
[0034] In order to construct a stable cell line with specific site integration, CHO-K1 cells were seeded into 6-well plates one day in advance, and when the cells expanded to 60%-80% fullness the next day, they were mixed with 500ng donor plasmid and 500ng CRI...
Embodiment 2
[0045] Example 2. Construction of CHO cell lines for protein display
[0046] In the present invention, a replacement expression frame is inserted into a specific site in the CHO cell genome, thereby constructing a high-expression cell line of a single-copy gene. This cell line can quickly replace any antibody gene or other genes into the genome, realize high transcription and expression of antibody genes and display antibodies on the surface of CHO cell membranes for affinity maturation of antibodies.
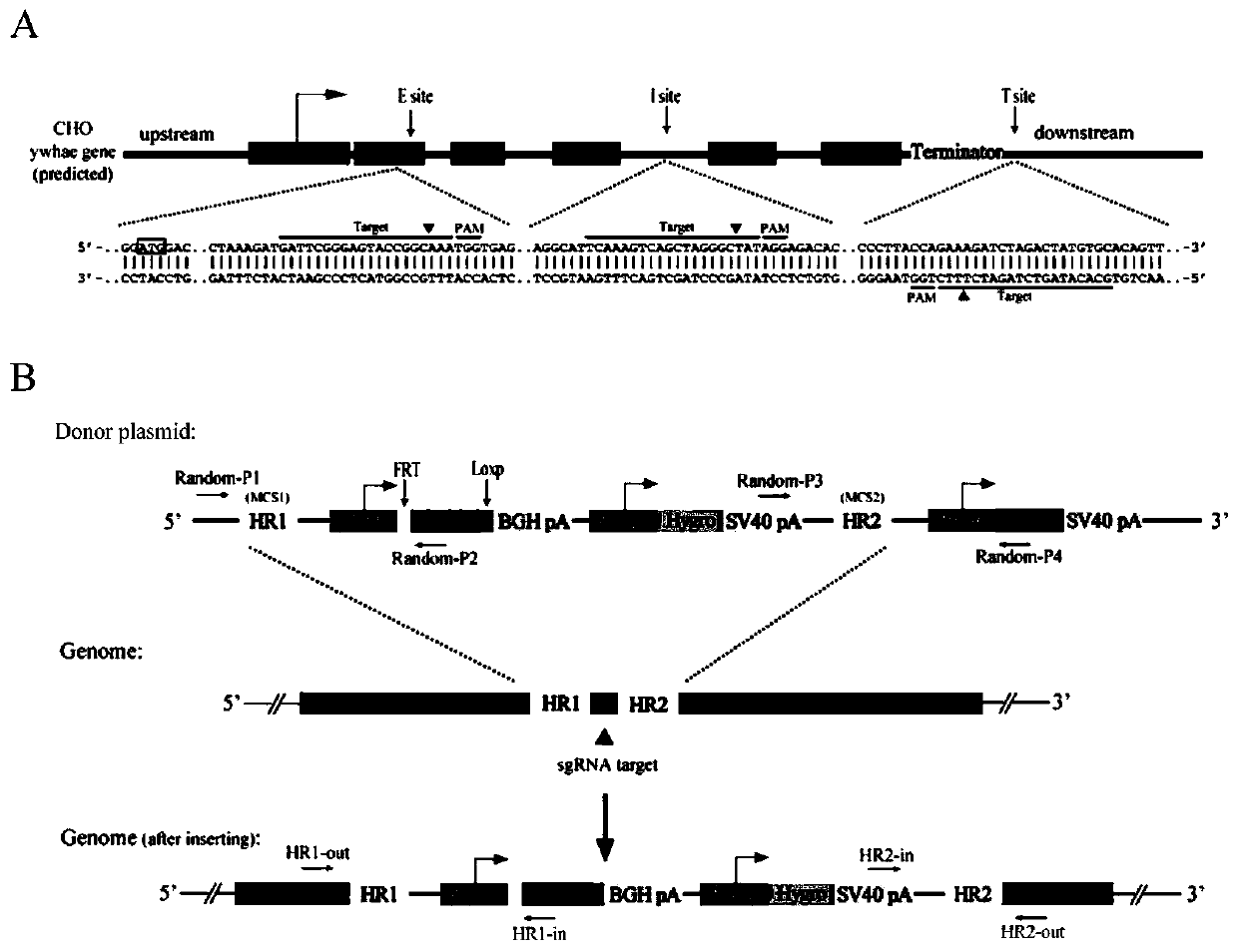
[0047] In the present invention, the YWHAE gene of the CHO cell genome is selected as the site-specific insertion position of the alternative expression cassette, and three sites of the YWHAE gene are selected for gene-site insertion of the alternative expression cassette. These three sites are E site (exon1), I site (intron, between exon 3 and exon 4) and T site (behind the stop codon of YWHAE gene), such as figure 1 shown.
[0048] The inserted alternative expression cassett...
Embodiment 3
[0052] Example 3, double-combinase-mediated target gene frame replacement
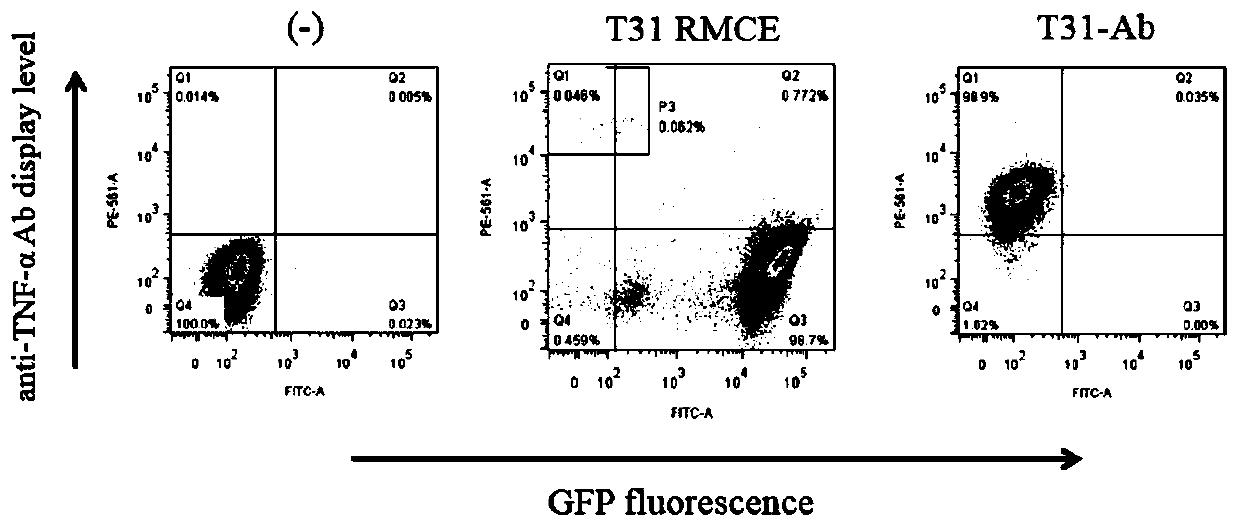
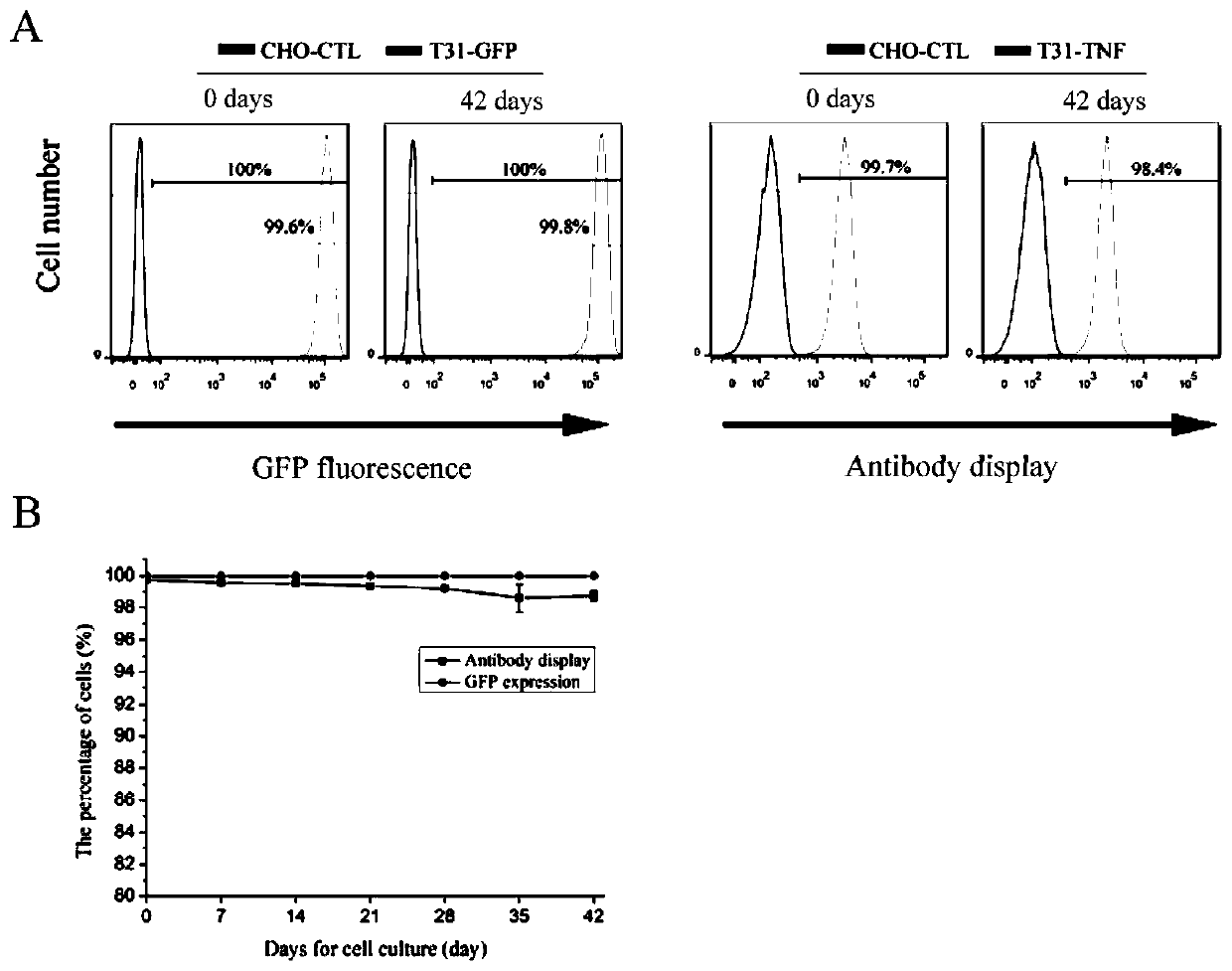
[0053] In order to detect whether the obtained CHO-K1-T cell line can perform double histone-mediated frame replacement and the corresponding protein expression (in the following embodiment, the CHO-K1-T cell line can also be used T31 or replace). We replaced the GFP gene in the clone with the RFP gene, anti-TNF-αScFv antibody gene and anti-HMGB1 ScFv antibody gene by RMCE. Cells expressing RFP or displaying antibodies were then sorted by flow cytometry. The sorted cells were cultured and expanded, and collected for flow analysis. The representative results are as follows: figure 2 shown.
[0054] Such as figure 2 As shown, the GFP gene in CHO-K1-T can be successfully replaced with the target gene, and the corresponding protein is expressed or displayed without continuing to express the GFP gene. The genome of the successfully replaced cells was extracted and primers (RMCE-P1 / P2) were designed t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com