High-throughput single-cell genome 5-hydroxymethylpyrimidine single-molecule visual analysis method
A technology of hydroxymethylpyrimidine single and hydroxymethylcytosine, which is applied in the field of high-throughput single-cell genome 5-hydroxymethylpyrimidine single-molecule visualization analysis, achieving the effect of fast reaction speed, simple reaction conditions and high yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
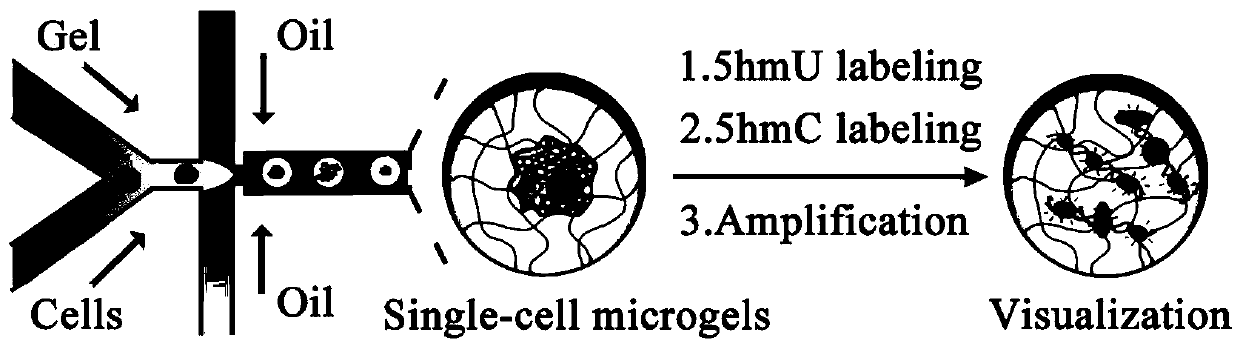
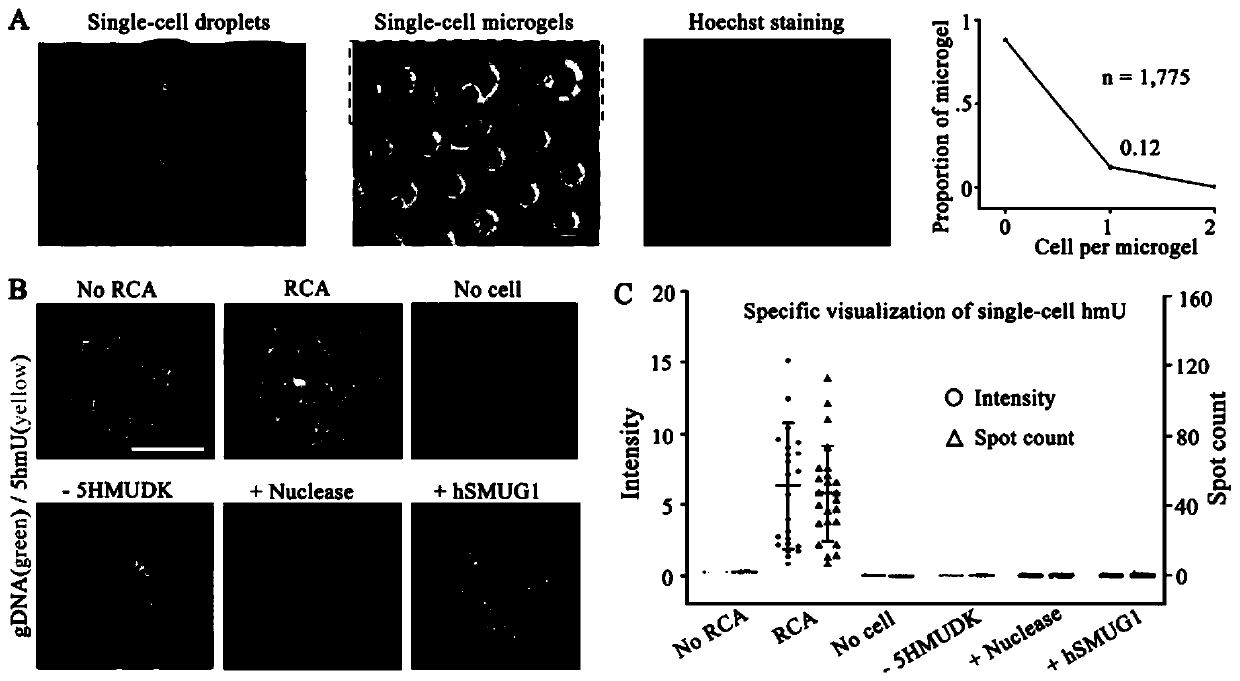
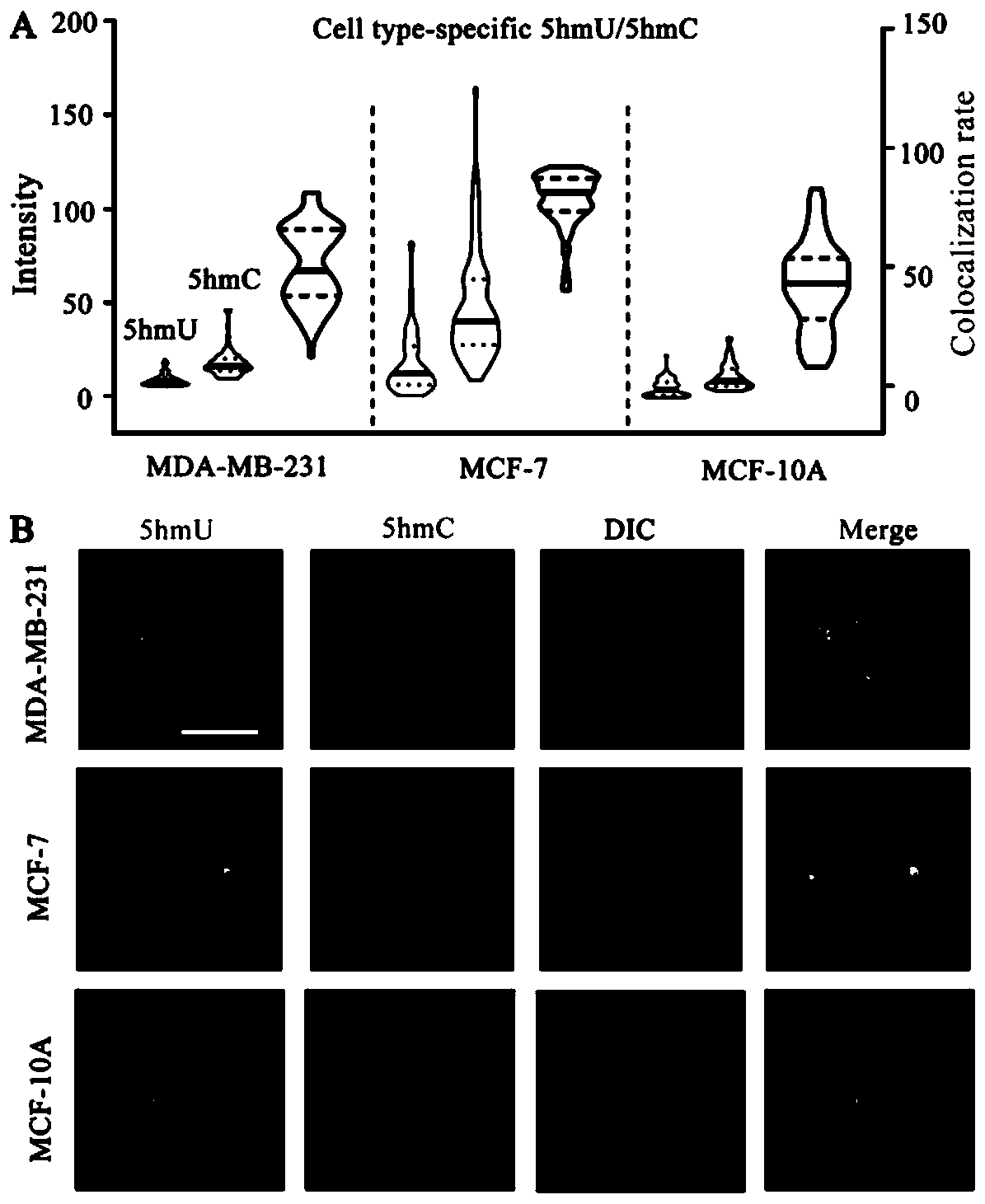
[0032] MCF-7 adherent cells were digested and a suspension of a certain cell density was prepared; a large number of agarose gel microspheres wrapped with single cells were obtained through a microfluidic chip, and after the single cells were lysed, each agarose coagulated Single-cell genomes are encapsulated in the gel microspheres. Then, primers were labeled with 5-hydroxymethylcytosine (5-hmC) and 5-hydroxymethyluracil (5-hmU) of the single-cell genome through specific chemical reactions, and then rolled circle amplification (RCA) And hybridize fluorescent probes, use SYBR Green I to stain the genome in the gel microspheres, and finally use fluorescent confocal microscopy for imaging analysis.
[0033] The number of 5-hmU and 5-hmC sites in a single cell can be assessed by the number of spots in each gel microsphere, enabling single-molecule visual analysis of the modified base 5-hmC and 5-hmU in a single cell .
[0034] The result is as figure 2 with image 3As shown,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com