Fluorescent signal enhancement method for detecting escherichia coli by enzyme substrate process
A technology of Escherichia coli and fluorescence signal, which is applied in the fields of biochemical equipment and methods, fluorescence/phosphorescence, and microbial determination/inspection, etc., can solve the problems of less 4-MU, inability to separate product signal and substrate signal, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Step 1. Purchase commercially available medium containing MUG enzyme substrate, 1L medium contains the following reagent components:
[0027] 4-Methylumbelliferone-β-D-glucuronide (MUG) 75.0mg
[0028] Tryptone 10.0g
[0029] Ammonium sulfate [(NH 4 ) 2 SO 4 ] 5.0g
[0030] Manganese sulfate (MnSO 4 ) 0.5mg
[0031] Zinc sulfate (ZnSO 4 ) 0.5mg
[0032] Magnesium Sulfate (MgSO 4 ) 100.0mg
[0033] Sodium chloride (NaCl) 10.0g
[0034] Calcium Chloride (CaCl 2 ) 50.0mg
[0035] Sodium sulfite (Na 2 SO 3 ) 40.0mg
[0036] K H 2 PO 4 0.9g
[0037] Na 2 HPO 4 6.2g
[0038] Step 2. Inoculate E.coli ATCC 25922 in LB medium by aseptic operation, cultivate to the stationary phase, and it takes about 12 hours at 37°C and 220 rpm. The cultivated E.coli was centrifuged and washed 3 times, and the OD was quantified 600 =0.2 is the bacterial solution A, and the bacterial solution A is diluted by 10 5 For bacterial solution B. Inoculate 50 μL of bacterial s...
Embodiment 2
[0044] Step 1. Purchase commercially available NA-MUG medium, and prepare the solution according to the instructions, 1L content:
[0045] Beef Extract 3.0g
[0046] Tryptone 5.0g
[0047] MUG 100mg
[0048] Step 2. is described with embodiment 1 step 2.
[0049] Step 3. Incubate the culture solution in step 2 at 44° C. for 12 hours.
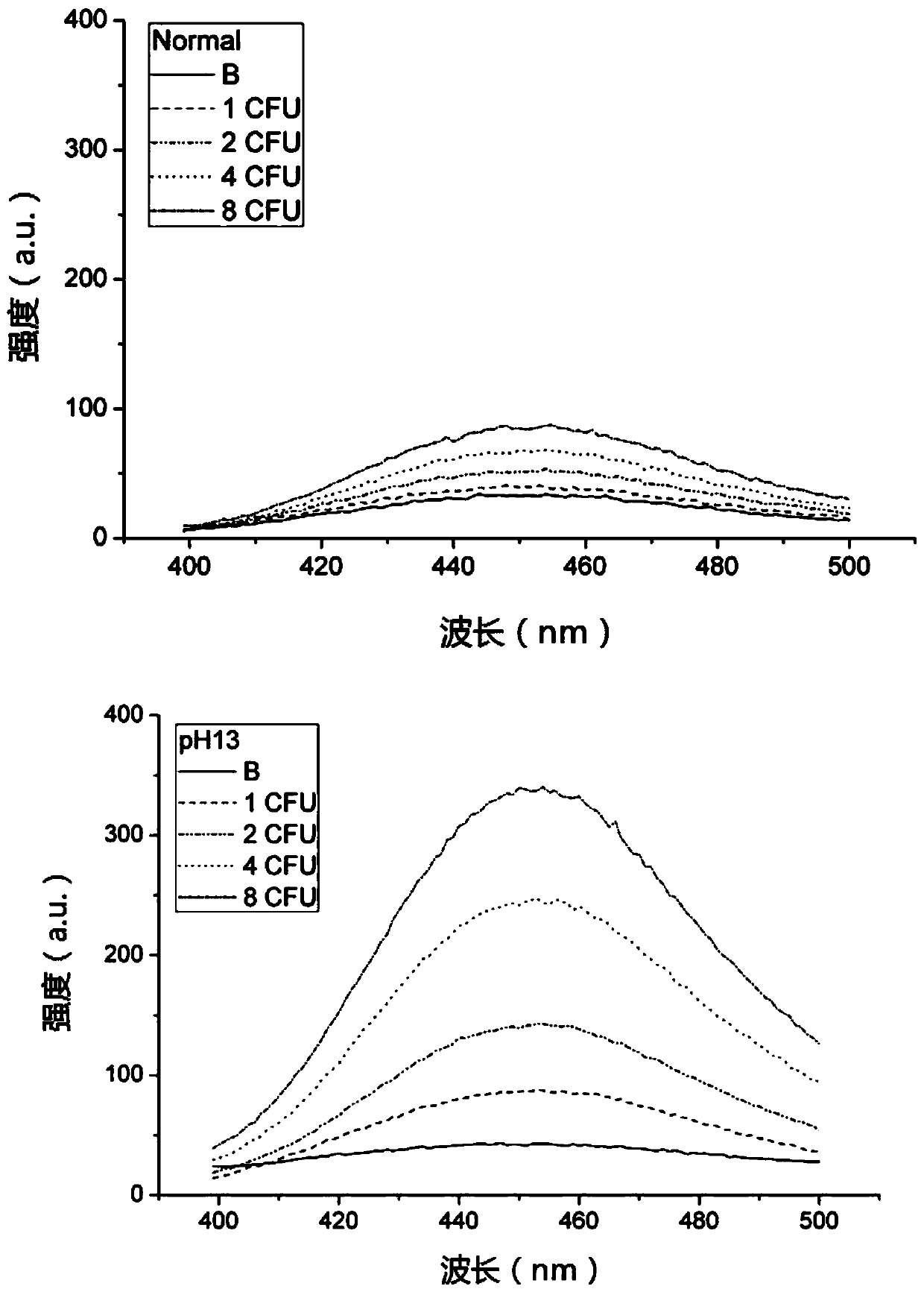
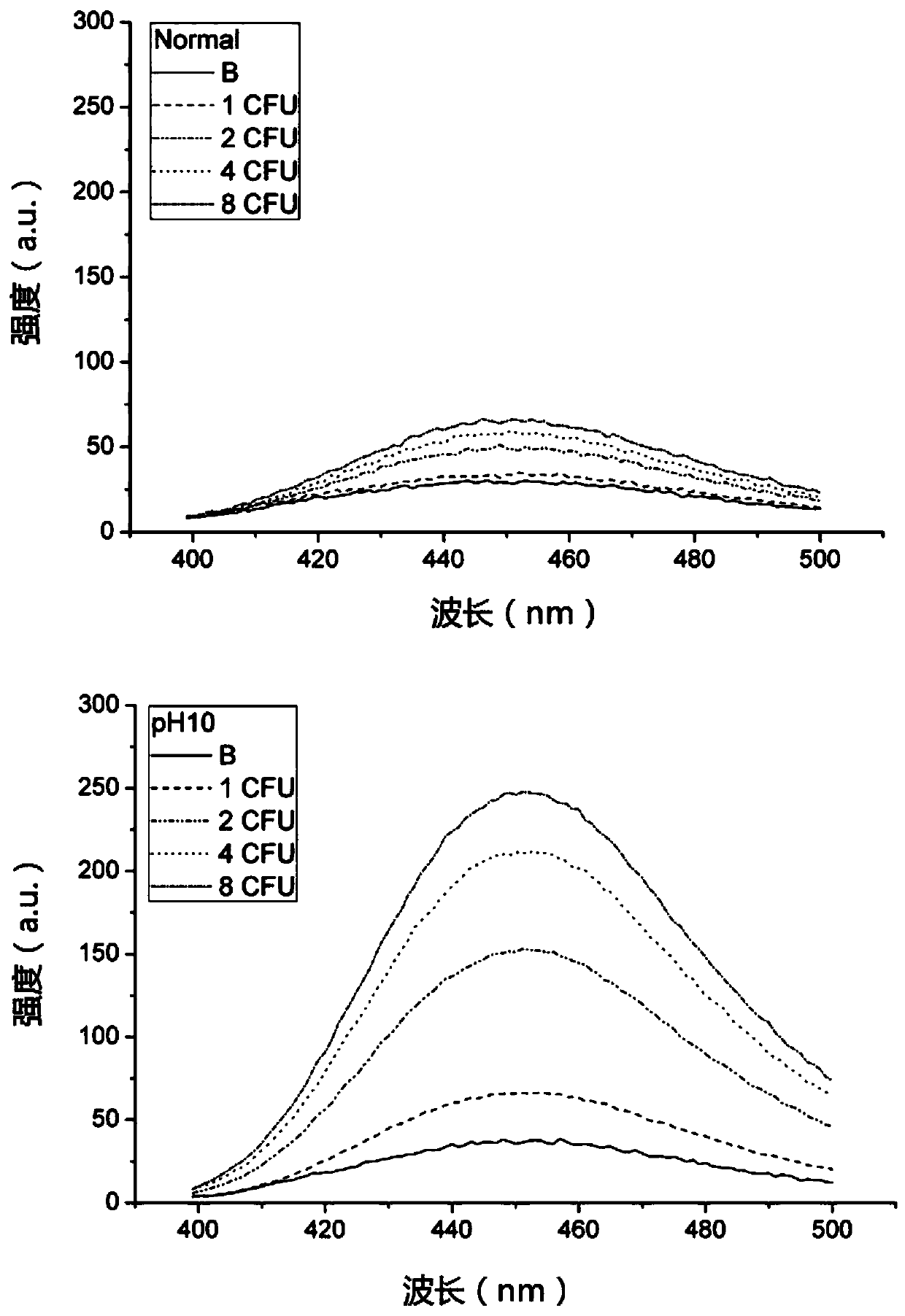
[0050] Step 4. Divide each tube of the solution in step 3 into two parts, one of which is adjusted to pH=10 by adding NaOH (marked as pH10), and the other part is used as a control (marked as Normal) for normal fluorescence measurement.
[0051] Step 5. Measure the fluorescence intensity of the two solutions described in step 4. Excited at 366nm, read the emission peak at 450nm.
[0052] Step 6. Results see figure 2 : It can be seen from the fluorescence measurement results of the two solutions marked as Normal and pH10 that the value of the fluorescence intensity subtracted from the blank of the same concentration sample increases by nea...
Embodiment 3
[0054] Step 1. Purchase commercially available MMO-MUG medium, and prepare the solution according to the instructions, 1L content:
[0055] Ammonium sulfate 5.0g
[0056] Manganese sulfate 0.5mg
[0057] Zinc sulfate 0.5mg
[0058] Magnesium Sulfate 100mg
[0060] Calcium chloride 50.0mg
[0061] Sodium sulfite 40.0mg
[0062] ONPG 500mg
[0063] MUG 75mg
[0064] HEPES 6.9g
[0066] Amphotericin B 1.0mg
[0067] Solanium Extract 500mg
[0068] Step 2. is described with the step 2 of embodiment 1.
[0069] Step 3. Incubate the culture solution in step 2 at 35° C. for 26 hours.
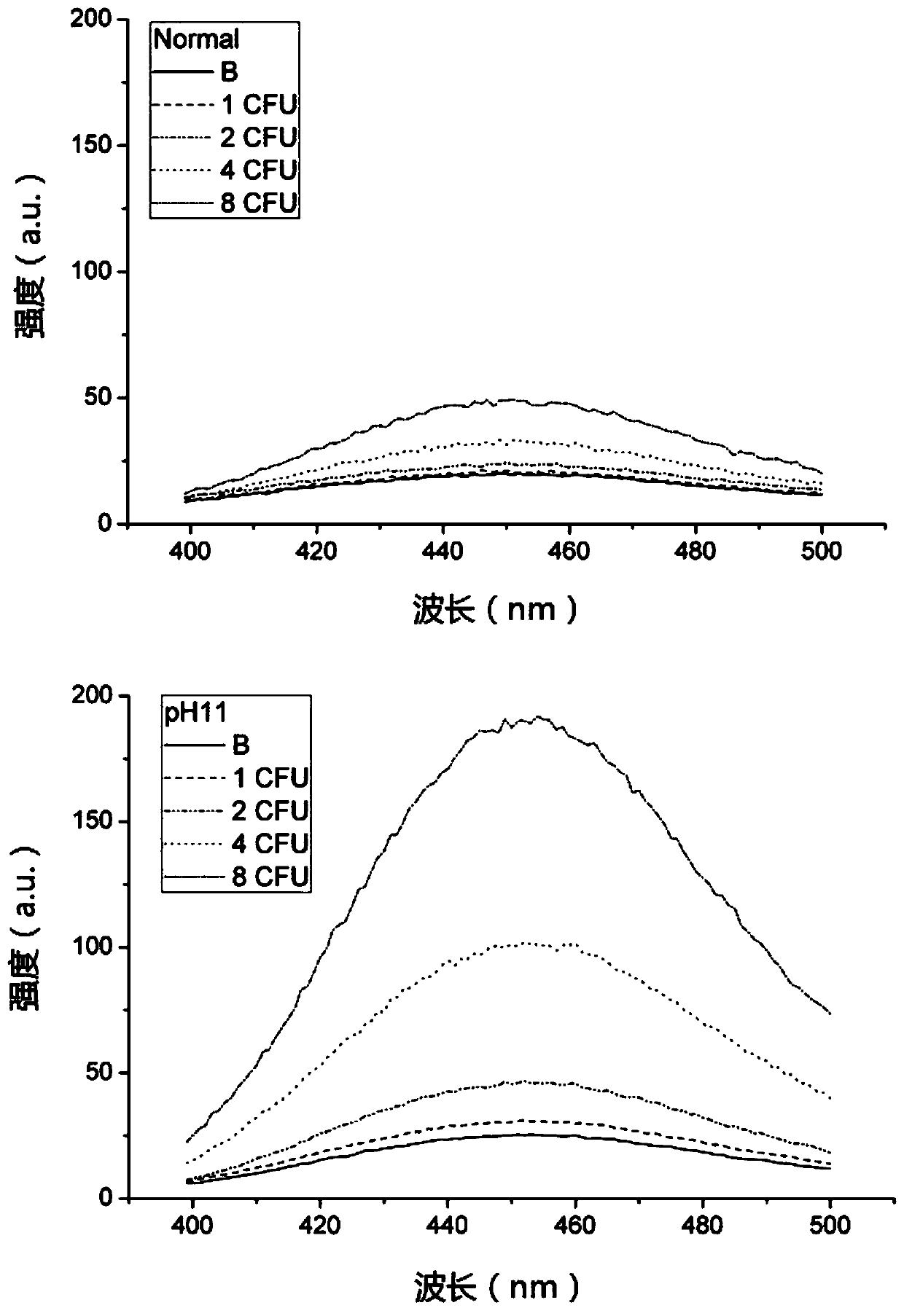
[0070] Step 4. Divide each tube of the solution in step 3 into two parts, one of which is adjusted to pH=11 by adding KOH (marked as pH11), and the other part is used as a control (marked as Normal) for normal fluorescence measurement.
[0071] Step 5. Measure the fluorescence intensity of the two solutions described in step 4. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com