Triple fluorescent quantitative PCR detection material and kit for distinguishing African swine fever virus wild strains from CD2V and/or 360-505R gene deleted strains
A fluorescence quantitative and gene deletion technology, applied in the field of virus strain identification, can solve the problem of inability to distinguish the infection of wild virus strains with gene deletion strains, and achieve the effects of saving detection cost and time, high sensitivity, and convenient clinical application and operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Embodiment 1 Primer and probe design
[0067] The inventor compared the P72, CD2V and 360-505R genes of African swine fever virus (ASFV) published in the GenBanK database of NCBI (National Center for Bioinformatics of the United States) in a large number of highly conserved and specific regions, and designed ASFV P72, CD2V and 360 The specific primers and probes for the -505R gene correspond to the three groups A, B, and C respectively. The specific sequences are:
[0068] Group A:
[0069] Upstream primer ASFV-P72-F:
[0070] TTTAATGAGAACGTGAACCTTGCT (SEQ ID NO: 1);
[0071] Downstream primer ASFV-P72-R:
[0072] ATGGTTTAAAGCGTATATTGCGTCT (SEQ ID NO: 2);
[0073] Specific fluorescent probe ASFV-P72-P:
[0074] Fluorophore - TCCCTCAGTATCCATTCCCTTCGGCGAG - Quencher (SEQ ID NO: 3);
[0075] Group B:
[0076] Upstream primer ASFV-CD2V-F:
[0077] CCTAAGCCTTACAGTCGTTATCAG (SEQ ID NO: 4);
[0078] Downstream primer ASFV-CD2V-R:
[0079] ACATGGTTTAGGTGGAGGACAT (SEQ ...
Embodiment 2 3
[0090] Embodiment 2 Triple fluorescent quantitative PCR detection method
[0091] Materials and Methods
[0092] 1.1 Primers in Example 1
[0093] 1.2 Sample DNA extraction
[0094] There are no special requirements for DNA extraction, which can be extracted according to conventional methods or DNA extraction kits. The extracted DNA was stored at -20°C for later use or immediately used for PCR amplification.
[0095] 1.3 Positive plasmid
[0096] Connect the PCR amplification products of three sets of primers A, B, and C to the pJET1.2 cloning vector, transform Escherichia coli competent cells DH5α, spread on LB medium plates containing 100mg / L ampicillin, and culture at 37°C After 12-16 hours, after picking bacteria, screening and sequencing identification, the plasmids were extracted after expanding the culture of positive bacteria, and the positive plasmids were named pJET1.2-P72(qPCR), pJET1.2-CD2V(qPCR), pJET1.2-360 -505R (qPCR).
[0097] 1.4 Triple fluorescent quan...
Embodiment 3
[0118] Embodiment 3 sensitivity test
[0119] Dilute the positive sample concentration to 1×10 -3 ng / μL, 1×10 -2 ng / μL, 1×10 -1 ng / μL, 1×10 0 ng / μL, 1×10 1 ng / μL.
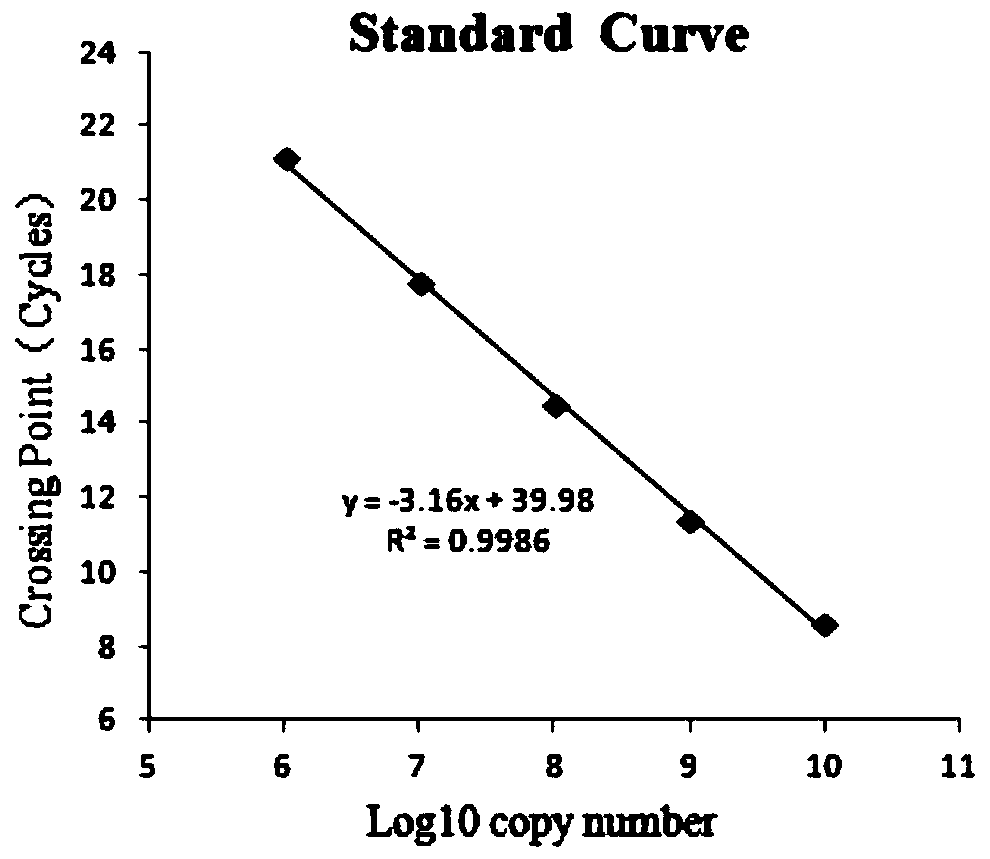
[0120] The above-mentioned different template concentrations were detected according to the triple fluorescent quantitative PCR detection method established in 1.4 of Example 2. The results are attached Figure 5 shown. From attached Figure 5 As can be seen in the 1×10 -3 The sample at the concentration of ng / μL still had a good amplification curve, indicating that the sensitivity of the method reached 1×10 -3 ng / μL.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com