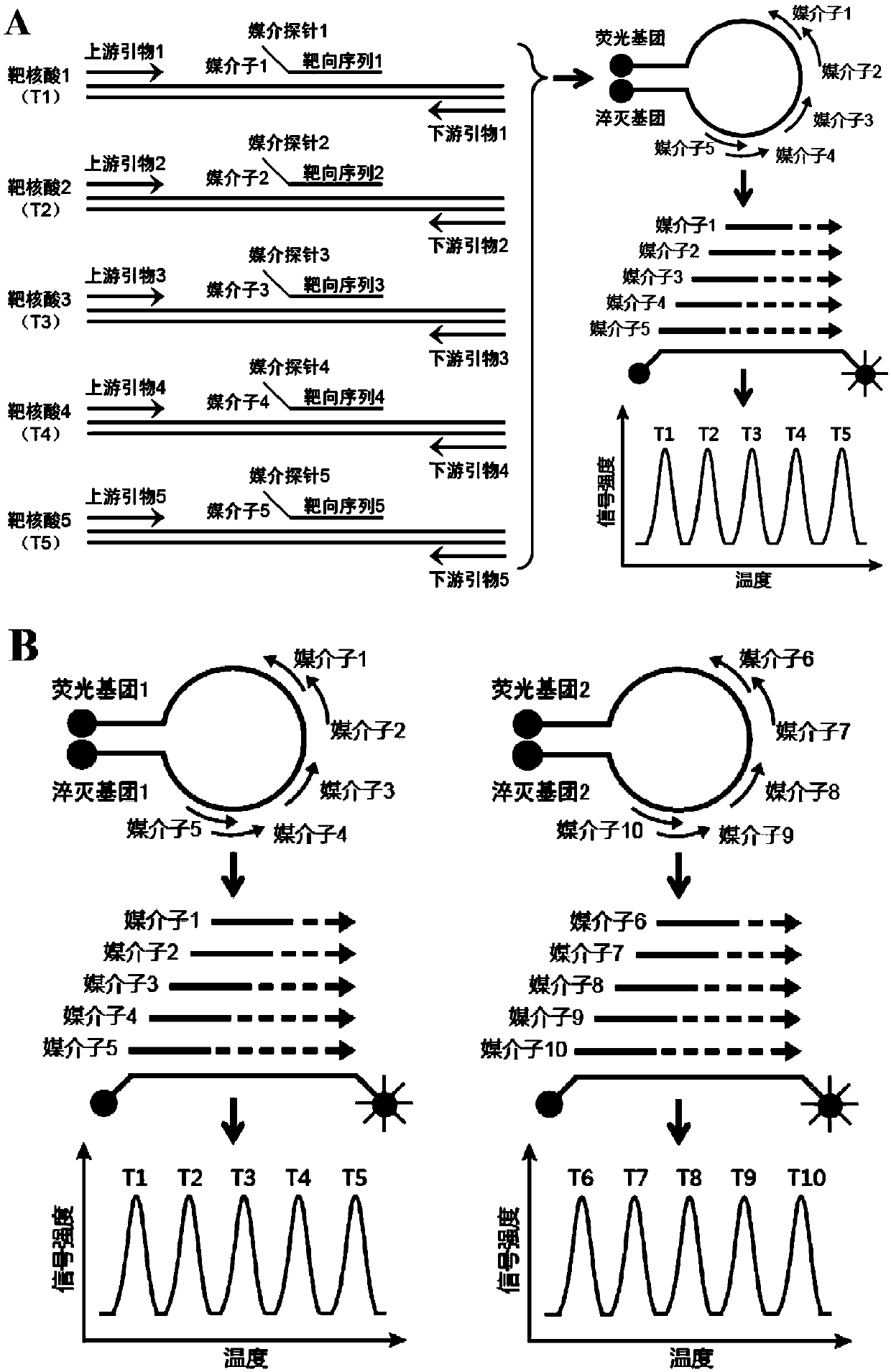
Detection method for antibiotic resistance genes
A technology for drug resistance genes and bacteria, which can be used in biochemical equipment and methods, determination/inspection of microorganisms, DNA/RNA fragments, etc., and can solve problems such as high cost and complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
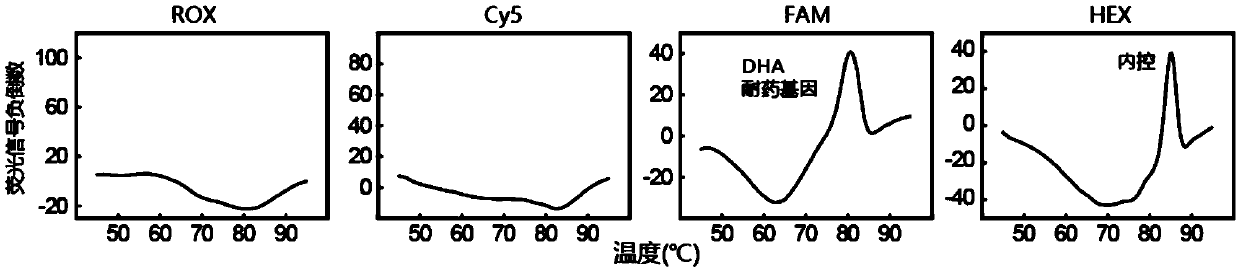
[0315] The detection of embodiment 1.DHA resistance gene
[0316] In this example, the reagents described in Table 1 (8 detection probes, 24 mediator probes, 24 upstream primers, 24 downstream primers, and 1 universal primer) and the detection protocol described in Table 2 were used , to detect samples containing DHA-resistant gene bacteria.
[0317] In short, in this example, a 25 μL PCR reaction system was used for real-time PCR, and the PCR reaction system included: 1×PCR buffer (67mM Tris-HCl, 50mM KCl and 0.085mg / mL BSA), 6.0mM MgCl 2 , 0.2mMdNTPs, 2.0U polymerase TaqHS (Takara), various reagents described in Table 1 (used at specified working concentrations), and 5 μL of bacterial DNA carrying DHA resistance gene and control DNA (Arabidopsis Lac8 gene) (the dosage ratio of the two is about 1:1). The reaction conditions of real-time PCR were: 95°C, 5min; then 50 cycles (95°C, 20s and 63°C, 1min). After PCR is finished, carry out melting curve analysis according to foll...
Embodiment 2
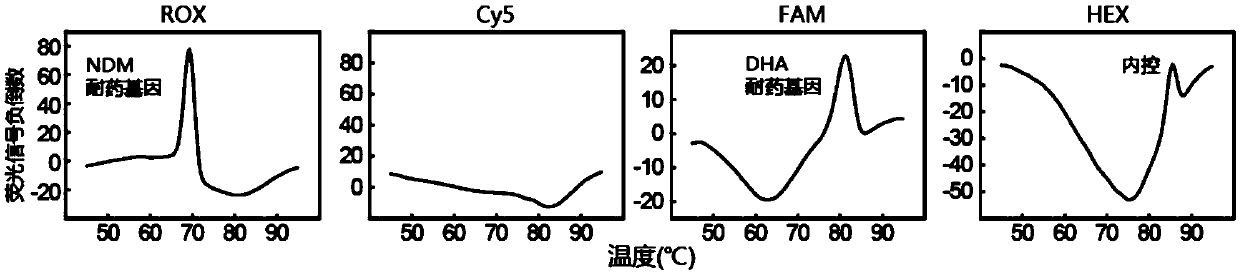
[0319] The detection of embodiment 2.NDM drug resistance gene and DHA drug resistance gene
[0320] In this example, the reagents described in Table 1 (8 detection probes, 24 mediator probes, 24 upstream primers, 24 downstream primers, and 1 universal primer) and the detection protocol described in Table 2 were used , to detect samples containing DHA-resistant gene bacteria and NDM-resistant gene bacteria.
[0321] In short, in this example, a 25 μL PCR reaction system was used for real-time PCR, and the PCR reaction system included: 1×PCR buffer (67mM Tris-HCl, 50mM KCl and 0.085mg / mL BSA), 6.0mM MgCl 2 , 0.2 mM dNTPs, 2.0 U of polymerase TaqHS (Takara), various reagents described in Table 1 (used at specified working concentrations), and 5 μL of bacterial DNA carrying NDM resistance gene, bacterial DNA of DHA resistance gene , and a mixture of control DNA (Arabidopsis Lac8 gene) (the amount ratio of the three is about 3:2:1). The reaction conditions of real-time PCR were: ...
Embodiment 3 24
[0323] Example 3. Twenty-four multiple detections
[0324] In this example, the reagents described in Table 1 (8 detection probes, 24 mediator probes, 24 upstream primers, 24 downstream primers, and 1 universal primer) and the detection protocol described in Table 2 were used , to contain 24 kinds of bacterial resistance genes (respectively from carrying SPM, GIM, NDM, IMP, VIM, SIM, CTX-M, OXA-51, GES, PER, OXA-58, ACT, CMY I, ACC, CMY II , DHA, FOX, OXA-24, KPC, OXA-23, OXA-48, VEB and SHV resistant gene bacteria) and control DNA (Arabidopsis Lac8 gene) samples were detected.
[0325] In short, in this example, a 25 μL PCR reaction system was used for real-time PCR, and the PCR reaction system included: 1×PCR buffer (67mM Tris-HCl, 50mM KCl and 0.085mg / mL BSA), 6.0mM MgCl 2, 0.2mMdNTPs, 2.0U of polymerase TaqHS (Takara), various reagents described in Table 1 (used at specified working concentrations), and 5 μL of nucleic acid mixture (which contains the DNA of the 23 drug-r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com