Efficient gene knockout vector and application thereof
A gene knockout and vector technology, applied in the field of gene editing, can solve problems such as unclear function and unclear copy number, and achieve the effect of reducing damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Construction of gene knockout vector
[0032]U6 promoter-driven gRNA cloning vector pX330 was purchased from Addgene (Plasmid No. 42230; Watertown, MA, USA). gRNAs targeting exon 1 of porcine pLeg1a (XM_003121211.1 / XP_003121259.1) and pLeg1b (XM_021074892.1 / XP_020930551.1) were designed using the online gRNA design tool (http: / / crispr.mit.edu).
[0033] Firstly, the gRNA recombination vectors of pX330-pLeg1a and pX330-pLeg1b were respectively constructed using the BpiI restriction site in pX330. The primers used for gRNA-pLeg1a are forward (SEQ ID NO:5): 5'-CACCGCATGGCTGGCAATATACTAC-3', reverse (SEQ ID NO:6): 5'-AAACGTAGTATATTGCCAGCCATGC-3'; and for gRNA-pLeg1b The primers were forward (SEQ ID NO:7): 5'-CACCGCTGGCATTACCTTGAGAGAC-3', reverse (SEQ ID NO:8): 5'-AAACGTCTCTCAAGGTAATGCCAGC-3'.
[0034] A primer containing U6- The 380 base pair fragment of gRNA-pLeg1b was then inserted into the XbaI site of the pX330-pLeg1a vector to generate a recombinant vec...
Embodiment 2
[0035] Example 2: Culture, transfection and screening of pig fetal fibroblasts
[0036] Porcine fetal fibroblasts (PFFs) were isolated from 32-day-old male Chinese experimental minipig embryos. These primary cells were cultured in high glucose Dulbecco's Medium Eagle medium (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; Gibco).
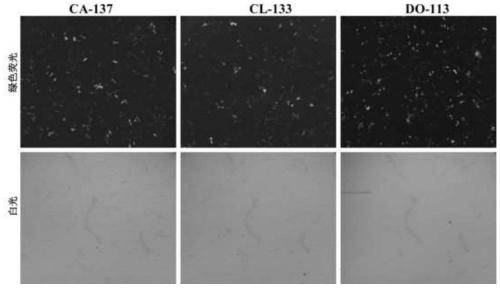
[0037] All animal experiments were performed according to the guidelines established by the China Animal Protection and Protocol Committee and were approved by the Experimental Animal Welfare Committee of Zhejiang University (Zhejiang Province, China). Transfection of PFF with pX330-pLeg1a-pLeg1b was performed using a Lonza Nucleofector. The day before transfection, PFFs were thawed and cultured. Then use 3 μg pMax (Lonza, Walkersville, MD, USA) to carry out transfection under different parameters (CA-137, CL-133 and DO-113), transfect about 1×10 6 PFF cells. The transfection effect was evaluated by the fluorescence intensi...
Embodiment 3
[0039] Example 3: Detection of Gene Modification and Integration of Exogenous Genes
[0040] Genomic DNA was extracted from each cell clone using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA), and then detected by PCR using high-fidelity Phusion polymerase (Thermo Fisher Scientific), and the pLeg1a-specific primers used were : 5'-CCCCCGCTGTGTGGAATAAGAG-3'(SEQ ID NO:11) and 5'-GCCCCACTGCAGTTTTTAGTAGC-3'(SEQ ID NO:12), the pLeg1b-specific primers used are: 5'-ACAAGGGAAGTGCCCTATTACC-3'(SEQ ID NO : 13) and 5'-TGATGTACAGGCAATCCCCA-3' (SEQ ID NO: 14).
[0041] PCR products were gel purified and then subjected to Sanger sequencing (BGI Genomics, Shenzhen, China).
[0042] In addition, the genomic DNA of the screened double gene knockout cell clones was also tested by PCR to verify the integration of the vector plasmid. The primers used were: Cas9-F (SEQ ID NO: 15): 5'-CATCGAGCAGATCAGCGAGT-3' and Cas9-R (SEQ ID NO: 16): 5'-CGATCCGTGTCTCGTACAGG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com