Isocitrate dehydrogenase kinase mutant and application thereof in preparation of aromatic amino acids
A technology for isocitric acid dehydrogenation and aromatic amino acids, applied in the field of biotechnology and molecular biology, to achieve the effect of clear genetic background and improved ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
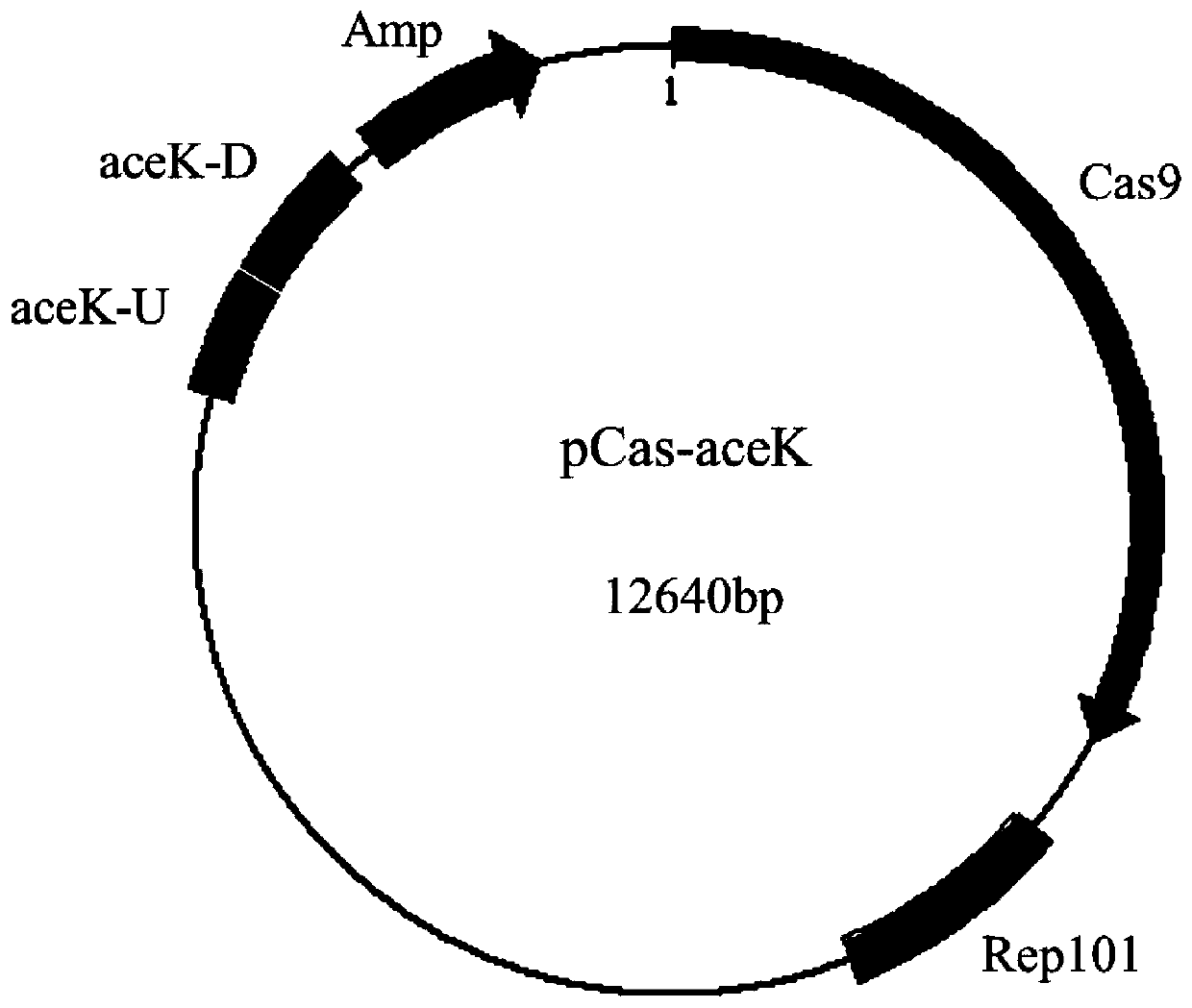
[0018] Example 1. Construction of pCas-aceK
[0019]The pCas-acek plasmid constructed in this example is used to replace the aceK wild-type gene on the genome of Escherichia coli tryptophan and phenylalanine engineering strains with the aceK mutant (E26K), and the specific working principle of the plasmid has been published ( Zhao D, Yuan S, Xiong B, Sun H, Ye L, Li J, Zhang X, Bi C. Development of a fast and easy method for Escherichia coli genome editing with CRISPR / Cas9..2016Dec 1;15(1):205 .)'. The specific construction process of pCas-red is described as follows: the Cas9 protein (as a plasmid containing the Cas9 protein (Addgene number: 42876)) was synthesized into a gRNA protein (sequence: 5'-GT T TTAGAGCTAGA A ATAGCAAGTTA AAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTT) that does not contain the N20 sequence -3') and its promoter sequence (5'-CTAGGTTTATACATAGGCGAGTACTCTGTTATGGAGTCAGATCT-3') and from pKD46 plasmid (Kirill A.Datsenko and Barry L.Wanner, One-step in...
Embodiment 2
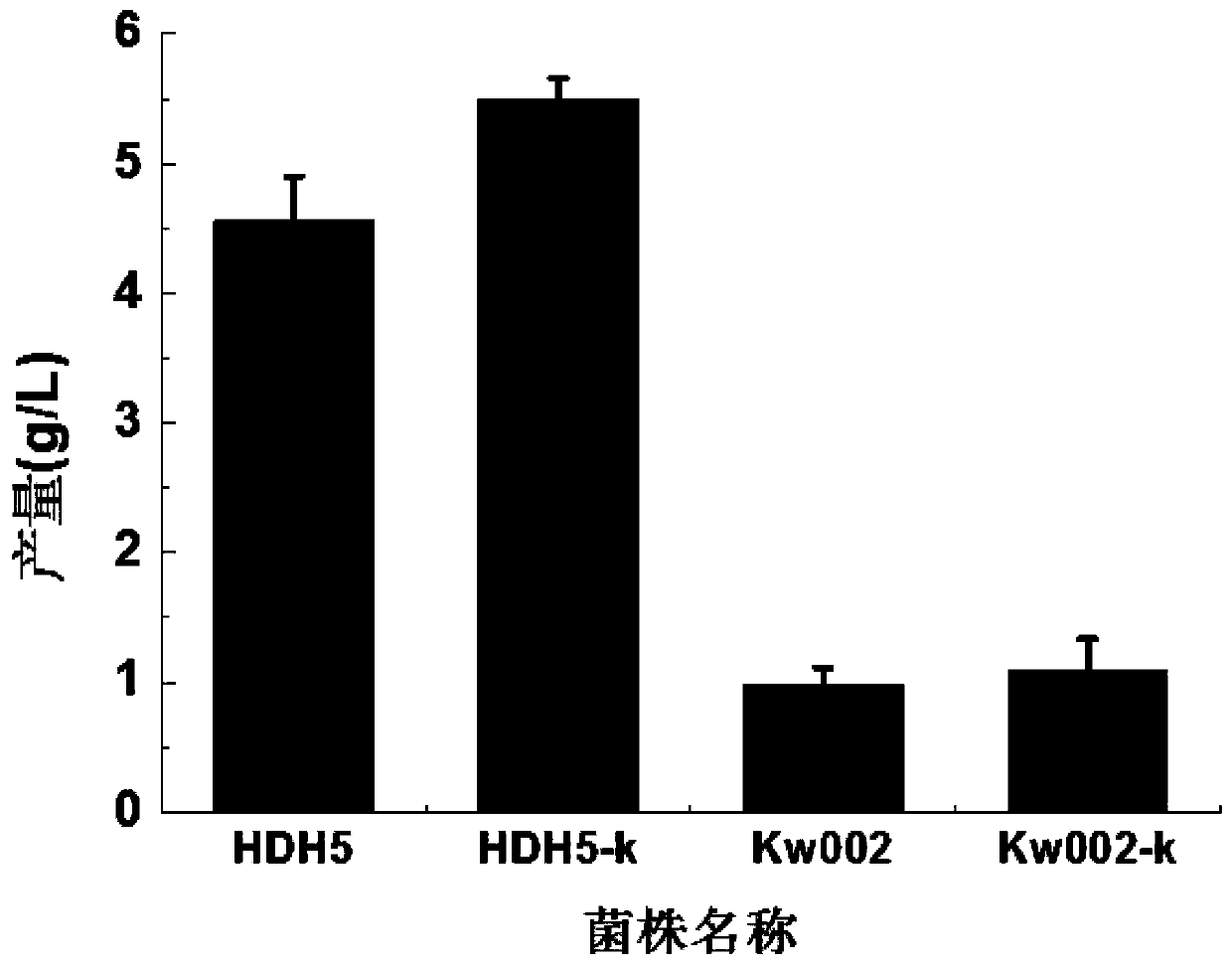
[0027] Example 2. Construction of HDH5-k
[0028] The Escherichia coli phenylalanine engineered strain HDH5 was prepared to be competent, and the pCas-aceK plasmid was transferred into the competent cells, and spread on a plate containing ampicillin resistance for overnight culture at 30°. Pick a positive single colony and inoculate it into a test tube containing ampicillin LB liquid medium (tryptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L), culture it under a shaker at 30°C for 6h, then add 2g / L Arabinose (induces the expression of Cas9 protein and recombinant protein on pKD46), promotes the selection of Cas9 protein. Then continue to culture at 30°C for 6 hours on a shaker, put the bacterial solution on the LB plate containing ampicillin and 2g / L arabinose for three-section line, and culture overnight at constant temperature at 30°C. Colony PCR was used for verification and sequencing, and the strains with correct sequencing were named HDH5-k.
Embodiment 3
[0029] Example 3. Construction of Kw002-k
[0030] The Escherichia coli phenylalanine engineered strain Kw002 was prepared to be competent, and the pCas-aceK plasmid was transferred into the competent cells, and then spread on a plate containing ampicillin resistance and cultivated overnight at 30°. Pick a positive single colony and inoculate it into an LB test tube containing ampicillin, culture it under a shaker at 30°C for 6 hours, and then add 2 g / L of arabinose (to induce the expression of Cas9 protein and PKD46 recombinant protein) to promote the screening of Cas9 protein. Then continue to culture at 30°C for 6 hours on a shaker, put the bacterial solution on the LB plate containing ampicillin and 2g / L arabinose for three-section line, and culture overnight at constant temperature at 30°C. Colony PCR was used for verification and sequencing, and the strains with correct sequencing were named Kw002-k.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com