Primer composition for detecting Y chromosome microdeletion and sex chromosome number and application
A technology of primer composition and chromosome number, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., and can solve the problems of high requirements for operators to interpret, long cycle, high cost of time and manpower, etc. , to achieve the effect of increasing the consistency of amplification and reducing the cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. Preparation of Y-del reaction solution and Y-del primer mixture
[0032] Table 1 Y-del reaction solution component preparation table
[0033]
[0034] Table 2 Y-del primer mixture components preparation table
[0035]
Embodiment 2
[0036] Embodiment 2 PCR amplification and result analysis
[0037] 1. Sample processing: Nucleic acid extractor (MagCore) and nucleic acid extraction kit (MagCore Genomic DNAWholeBlood Kit) extract human genomic DNA for subsequent PCR reaction, DNA concentration is 0.5ng / uL~50ng / uL, ratio of OD260nm / OD280nm Between 1.6-2.0.
[0038] 2. Preparation of amplification reagents:
[0039] (1) Take out the Y-del reaction solution and Y-del primer mixture from the kit, thaw at room temperature, mix them upside down, and centrifuge briefly with a microcentrifuge to make all the liquid settle to the bottom of the tube.
[0040] (2) Preparation of amplification reagents: prepare amplification reagents as shown in Table 3
[0041] Table 3 Amplification reagent preparation table
[0042]
[0043] * In order to reduce the separation error, it is recommended to take (n+1) parts of reaction solution and mixed enzyme according to the number of samples (n) when preparing amplification reage...
Embodiment 3
[0062] Example 3 Reagent Performance Verification
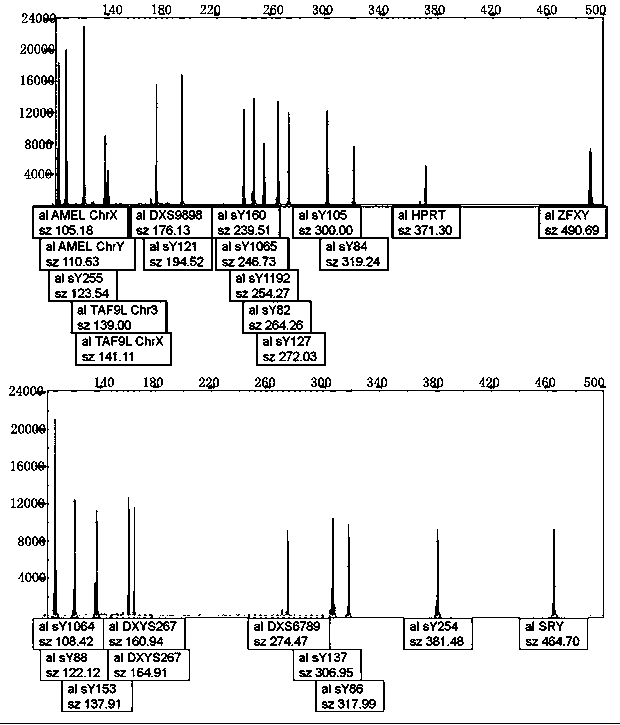
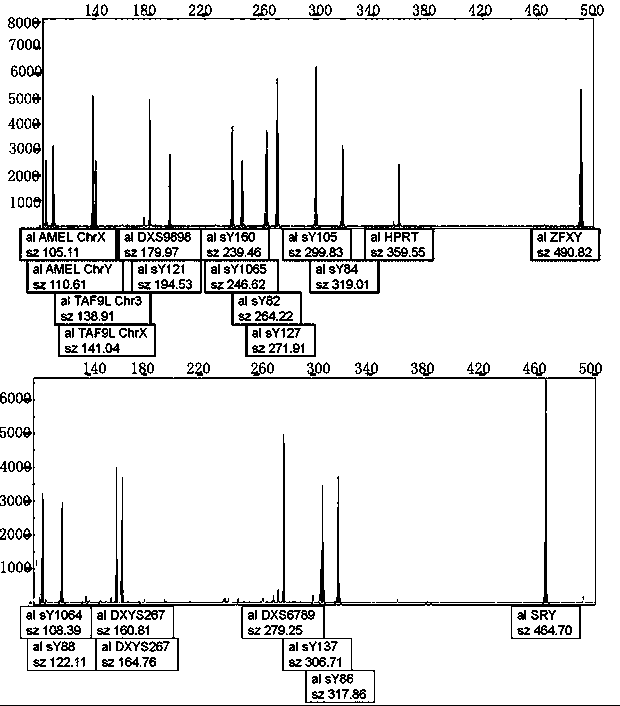
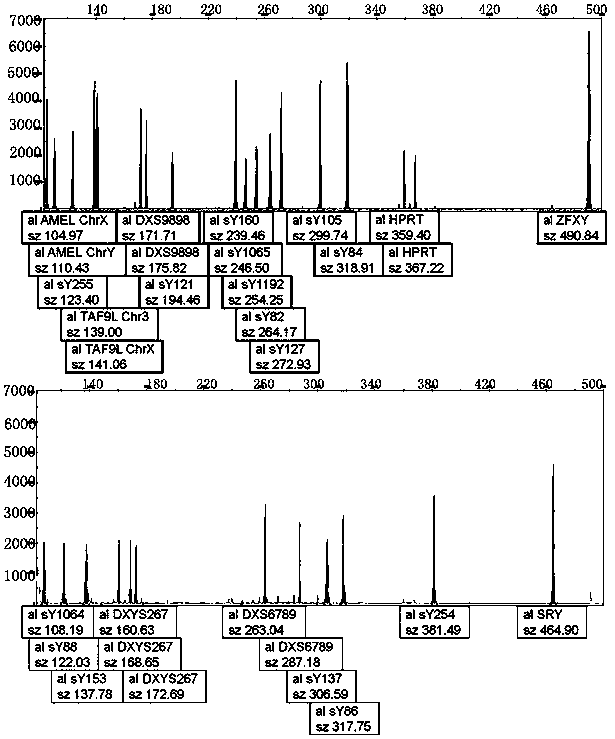
[0063] 1. The Y-del normal male control was prepared using conventional methods. According to the amplified sequence of each detection site, a plasmid was synthesized, which contained the detection sites SRY, ZFX / Y, sY84, sY86, sY82, sY1064, sY1065, sY88, sY127 , sY134, sY105, sY121, sY1192, sY153, sY254, sY255, sY160, AMEL, TAF9L, DXYS267, DXS9898, DXS6789, and HPRT were mixed in equal mass ratios, and the final concentration was 14,500 copies / μL.
[0064] 2. The preparation of Y-del normal female control adopts conventional methods. According to the amplified sequence of each detection site, the plasmid is synthesized, and the plasmid with the detection site ZFX / Y, AMEL, TAF9L, DXYS267, DXS9898, DXS6789, HPRT, etc. The mass ratio was mixed, and the final concentration was 14500 copies / μL.
[0065] 3. The coincidence rate of positive reference products was tested for 6 positive reference products. As shown in the table belo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com