Method for quantitatively co-expressing multiple proteins in vitro and application thereof
A co-expression and protein technology, applied in the field of quantitative co-expression of multiple proteins in vitro, to achieve the effect of simplification of complex phenomena and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0143] DNA Screening and Codon Optimization of Expression Sequences of Different Fluorescent Proteins
[0144] By searching different databases, Blast was performed on the coding sequences of 18 different fluorescent protein genes, and the codons were optimized to make them suitable for Kluyveromyces lactis in vitro cell-free protein synthesis system.
Embodiment 2
[0146] Construction of in vitro cell-free protein synthesis system plasmids containing eukaryotic cell translation regulatory sequences, fluorescent protein coding sequences, and tag (protein expression and purification sequences)
[0147] 2.1 Whole gene synthesis
[0148] The optimized DNA sequence in Table 2 was used for genome synthesis.
[0149] 2.2 Primer design
[0150] Primers were designed with oligo 7.0 software, and the primer sequences are shown in Table 2.
[0151] Table 2 Primer Sequence
[0152]
[0153]
[0154]
[0155] 2.3 Construction of plasmid
[0156] Insert the gene sequence of the target protein to be expressed into the pD2P plasmid (see Figure 11 ), the specific construction process is as follows:
[0157] Using molecular cloning technology, pD2P was used as a template for plasmid construction, and two pairs of primers were used for PCR amplification, and 8.5 μL of the two amplification products were mixed with 1 μL of DpnI and 2 μL of 10...
Embodiment 3
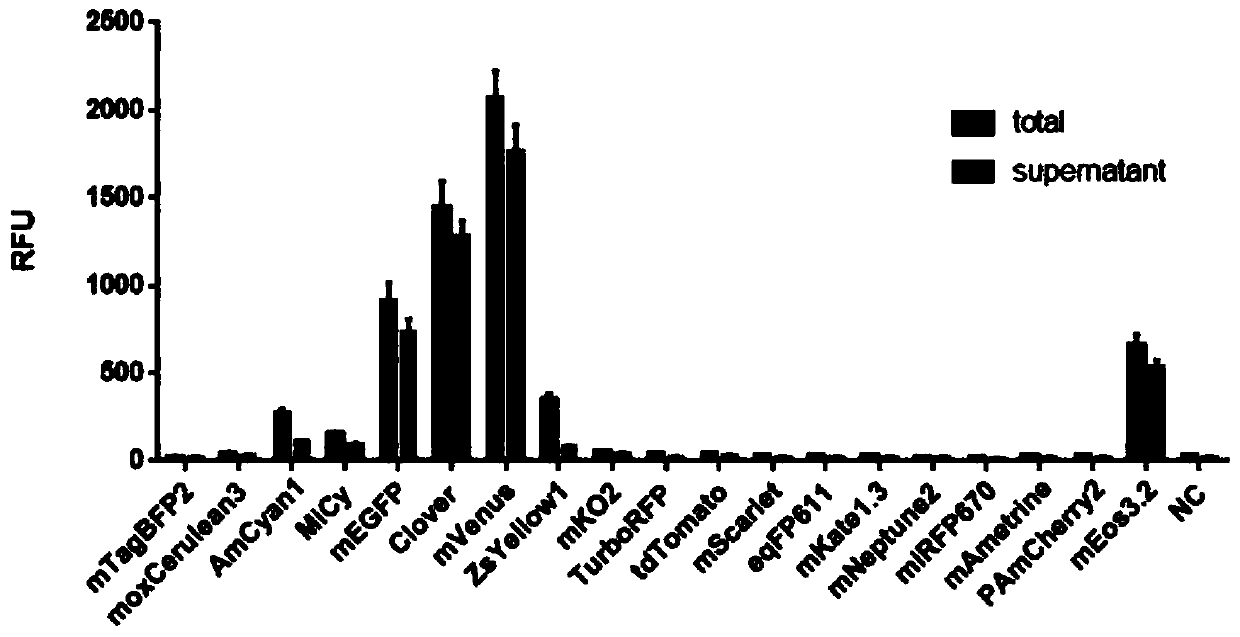
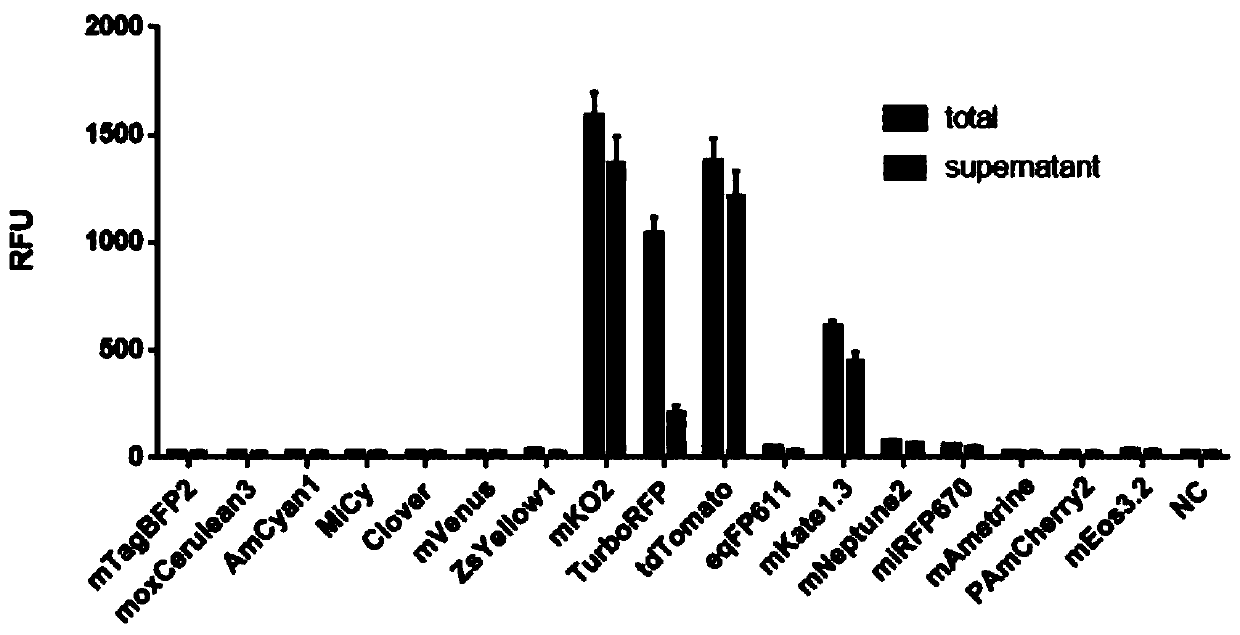
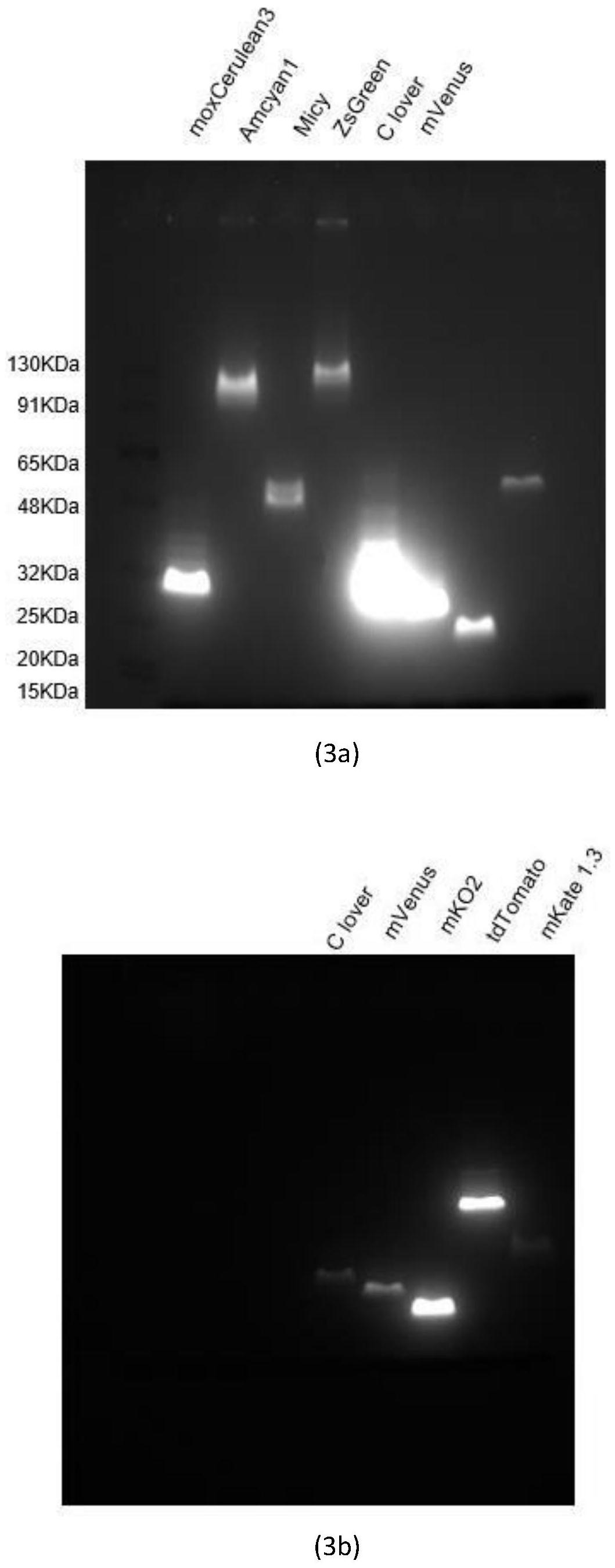
[0160] Expression of different fluorescent proteins by yeast in vitro cell-free protein synthesis system
[0161] Using the method of phi29 DNA polymerase amplification, using the above plasmids as templates, using random seven-base primers to amplify the fragments located between the 5'UTR of the transcription initiation sequence and the 3'UTR of the termination sequence in all plasmids, the amplified The product serves as a DNA template for the synthesis of various fluorescent proteins. Wherein, the transcription start sequence 5'UTR and the termination sequence 3'UTR contain one or more tandem combinations, and the tandem combinations include enhanced translation regulatory elements and protein expression purification tag elements.
[0162] According to the instructions for use, the fluorescent protein DNA template prepared above (the concentration of the template mother solution of different fluorescent proteins is equal) was added to the self-made Kluyveromyces lactis in ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com