Nucleic acid, kit and droplet type digital PCR method for detecting infectious bovine rhinotracheitis virus
A rhinotracheitis virus, infectious technology, applied in microorganism-based methods, biochemical equipment and methods, microorganisms, etc., to achieve the effects of good airtightness, high accuracy, and avoidance of pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1 The establishment of IBRV ddPCR method
[0037] The gB gene sequence of IBRV published by GenBank (if available, accession number: KU198480.1\JN787952.1\KY215944.1\MH751901.1\MG407790.1\KM258883.1\KM258880.1\KM258881.1\JK898220.1 etc.) carried out sequence comparison, and further determined the conserved region of the IBRV gB gene (the 1800-2100th DNA fragment, its sequence is as shown in the sequence of SEQ ID NO.4), and designed for the head, middle and tail of the conserved region Primers and probes were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.; the 5' end of the probe was labeled with a FAM fluorescent group, and the 3' end of the probe was labeled with a BHQ1 quencher group. The designed primers and probe sequences are described in part, and the specific sequences are shown in Table 1.
[0038] Table 1 The sequences of primers and probes for IBRV ddPCR detection
[0039]
[0040] * Indicates the position in the gB gene.
[0041] 2...
Embodiment 2I
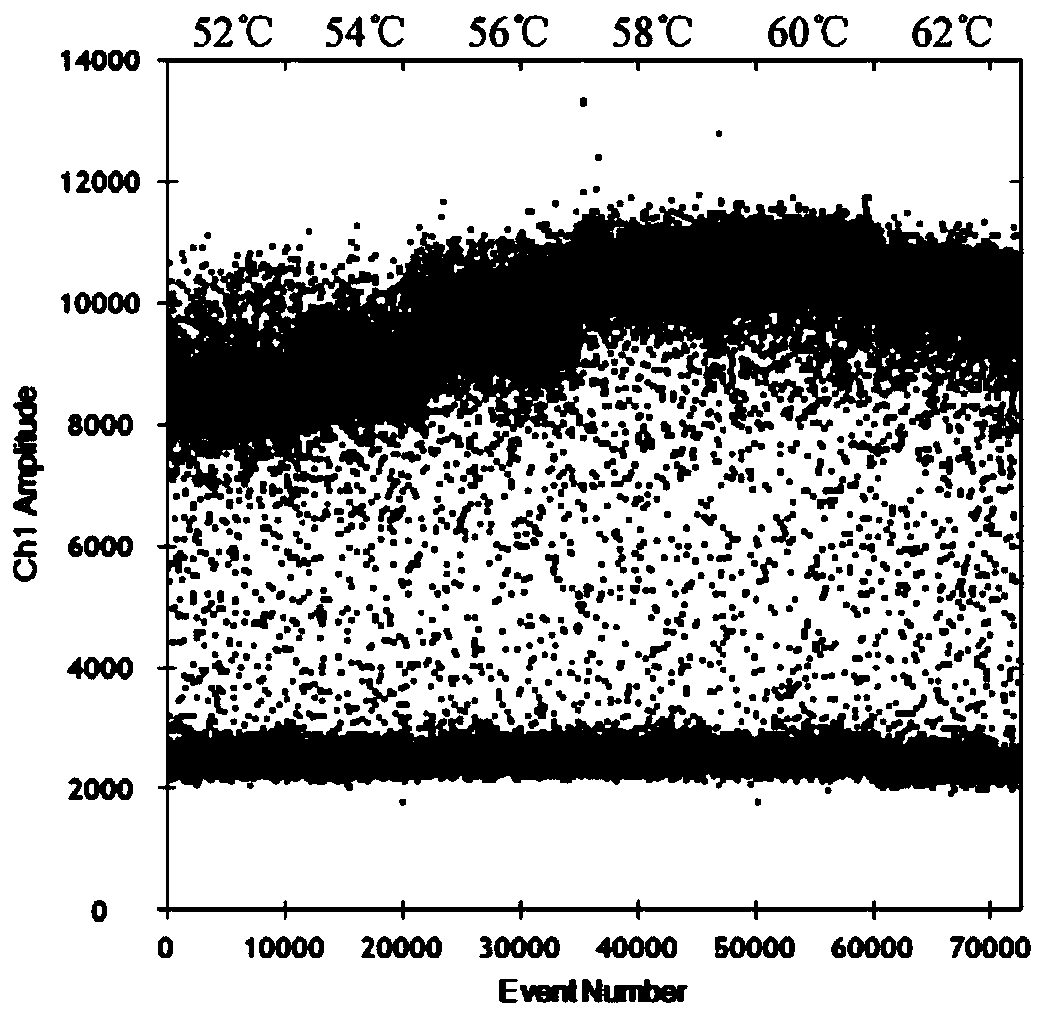
[0057] The optimization of embodiment 2 IBRV ddPCR annealing temperature
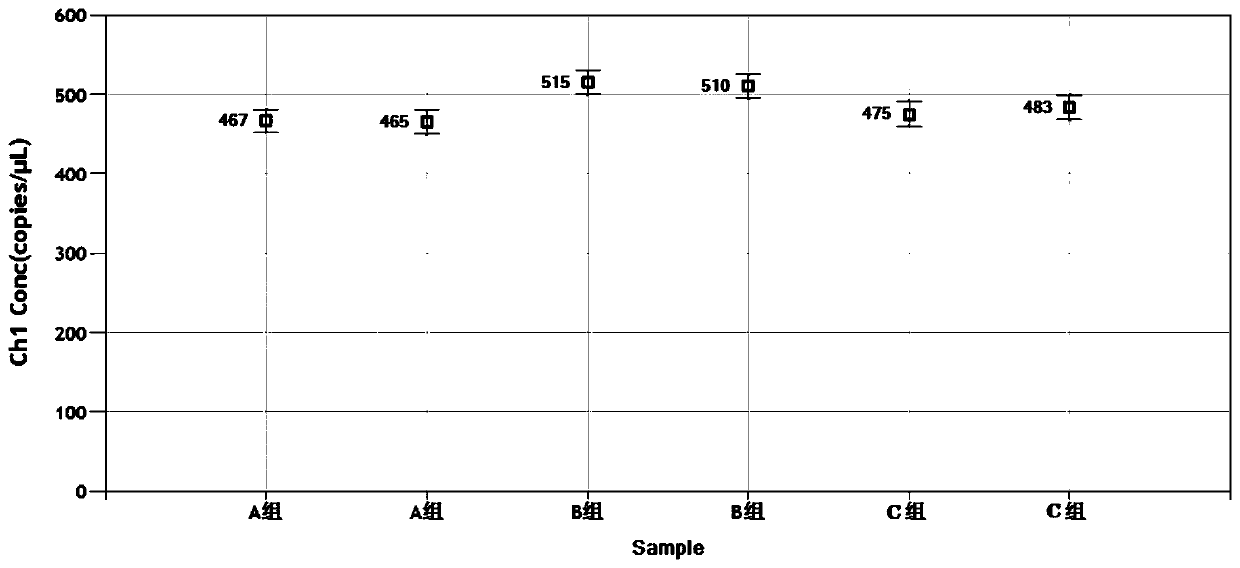
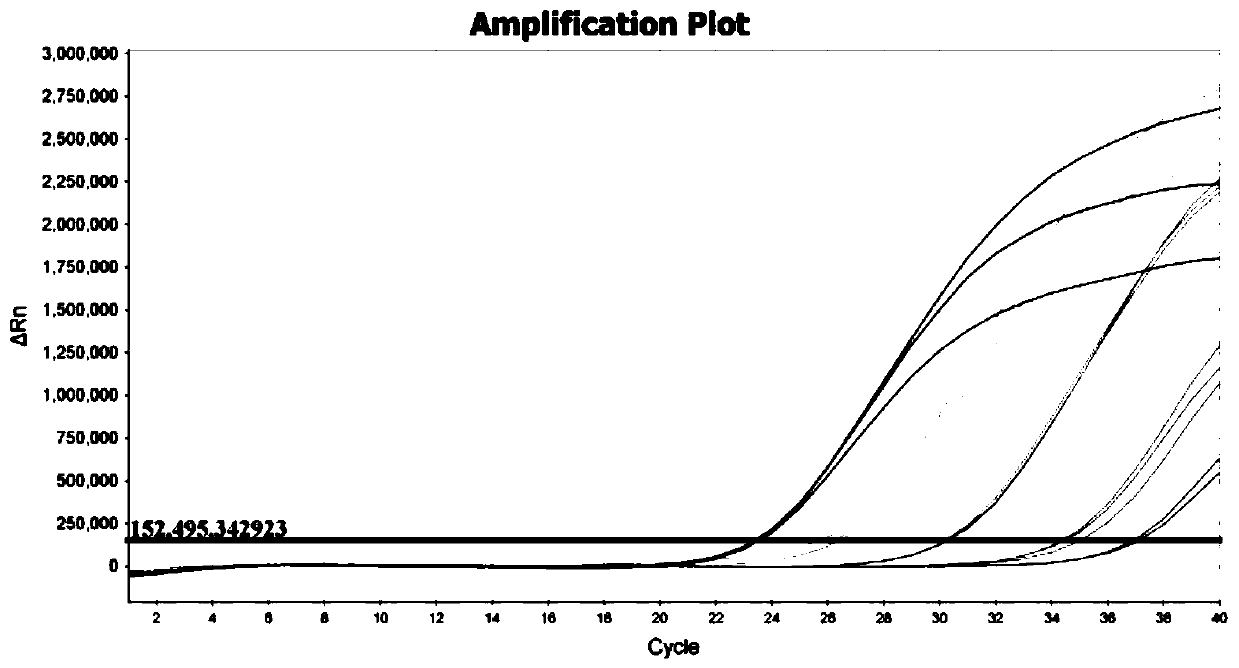
[0058] The synthetic IBRV gB gene DNA plasmid (pIBRV / ddPCR plasmid) in embodiment 1 is diluted to 1.0 * 10 5 Copies / μL is the template for ddPCR detection. The ddPCR reaction system includes 10 μL of 2×ddPCR Supermix, 2 μL of DNA template, 1.8 μL of forward primer (SEQ ID NO.1) (10 pmol / μL), reverse primer (SEQ ID NO. .2) (10 pmol / μL) 1.8 μL, probe (SEQ ID NO.3) (10 pmol / μL) 0.5 μL, DNase-free water 3.9 μL, the total volume is 20 μL.
[0059] Make 6 duplicate wells, transfer to the droplet generation card, and place it on the droplet generator to generate droplets. The generated micro-droplets were transferred to a 96-well reaction plate and sealed with a PX1 heat sealer at 180°C for 5 s. Place the sealed 96-well reaction plate on a PCR machine for amplification.
[0060] The reaction conditions are: pre-denaturation at 95°C for 10 minutes; denaturation at 94°C for 30 seconds, annealing at 52°C, 54°C...
Embodiment 3I
[0061] The determination of embodiment 3IBRV ddPCR primer concentration and probe concentration
[0062] The synthetic IBRV gB gene DNA plasmid (pIBRV / ddPCR plasmid) in embodiment 1 is diluted to 1.0 * 10 5 Copies / μL is the template for ddPCR amplification. The ddPCR reaction system includes 10 μL of 2×ddPCR Supermix forProbes, 2 μL of DNA template; the final concentration of forward primer (SEQ ID NO.1) and reverse primer (SEQ ID NO.2) 500-900nM, the final concentration of the probe (SEQ ID NO.3) is 100-300nM, the final concentration of the primer is increased by 100nM, and the final concentration of the probe is increased by 50nM for orthogonal test; the rest is made up to 20μL with DNase-free water .
[0063] The reaction parameters were: pre-denaturation at 95°C for 10 min; denaturation at 94°C for 30 s, annealing at 58°C for 30 s, 40 cycles; 10 min at 98°C; 4°C∞, heating and cooling rate 2°C / s.
[0064] The results show that the final concentration of forward primer and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com