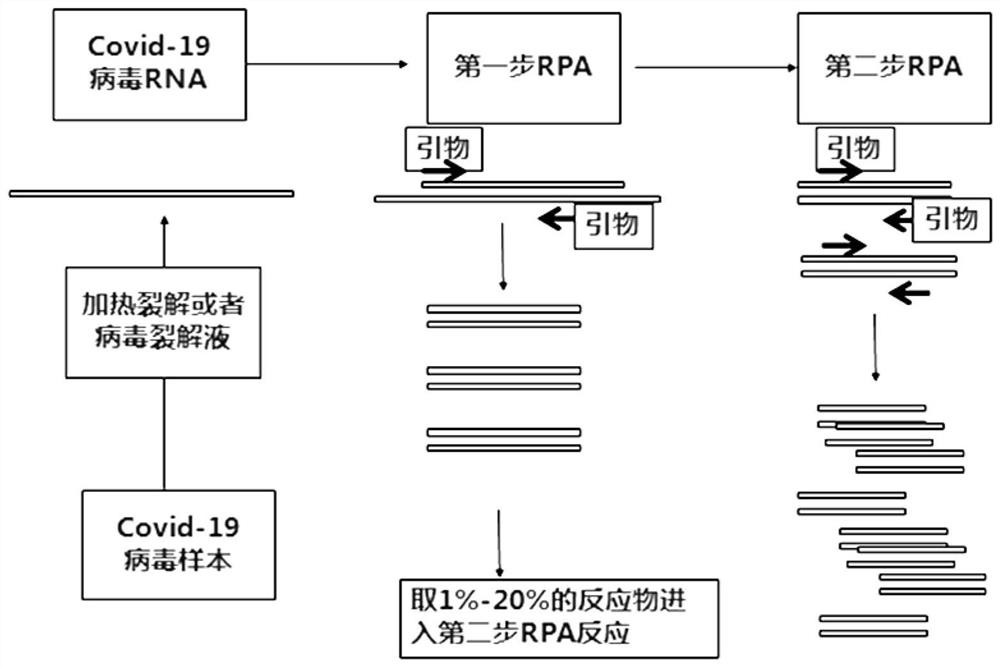
Virus recombinase-polymerase amplification detection method
A virus detection and virus technology, applied in the field of detection of new coronavirus, to achieve high sensitivity, speed up, and increase the effect of amplification multiple
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
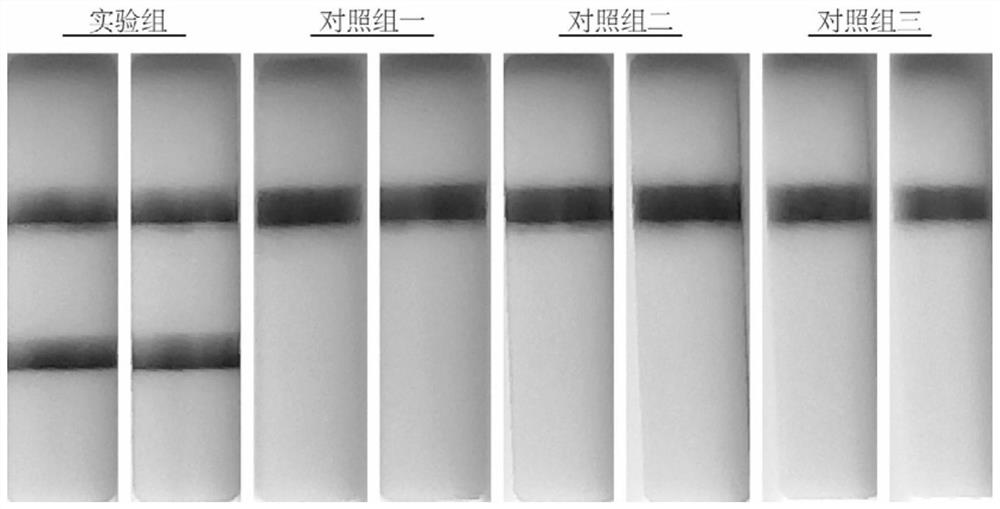
[0060] Example 1 Two-step RPA detection of COVID-19 virus RNA validity verification
[0061] 1. Reagent Preparation
[0062] COVID-19 virus RNA samples were purchased from national standard materials (standard material number: GBW(E)091099)
[0063] 1 st The RPA reaction kit was purchased from TwistDX Company in the UK, and the product model was Basic RT kit
[0064] 2 nd The RPA reaction kit was purchased from TwistDX Company in the UK, and the product model was nfo kit
[0065] Colloidal gold test paper was purchased from Beijing Kuer Technology Co., Ltd.
[0066] 2. Experimental steps
[0067] 1) The new coronavirus RNA stock solution (E gene RNA concentration: 1.06±0.11×10 3 copies / ul) diluted to 10copies / ul.
[0068] 2) Dilute the forward primer, reverse primer and probe to 10 uM with DEPC water. The primer sequences are as follows:
[0069] 1 st RPA-F: ATGTACTCATTCGTTTCGGAAGAGACAGG (SEQ ID No. 1)
[0070] 1 st RPA-R:AGACCAGAAGATCAGGAACTCTAGAAGAA (SEQ ...
Embodiment 2
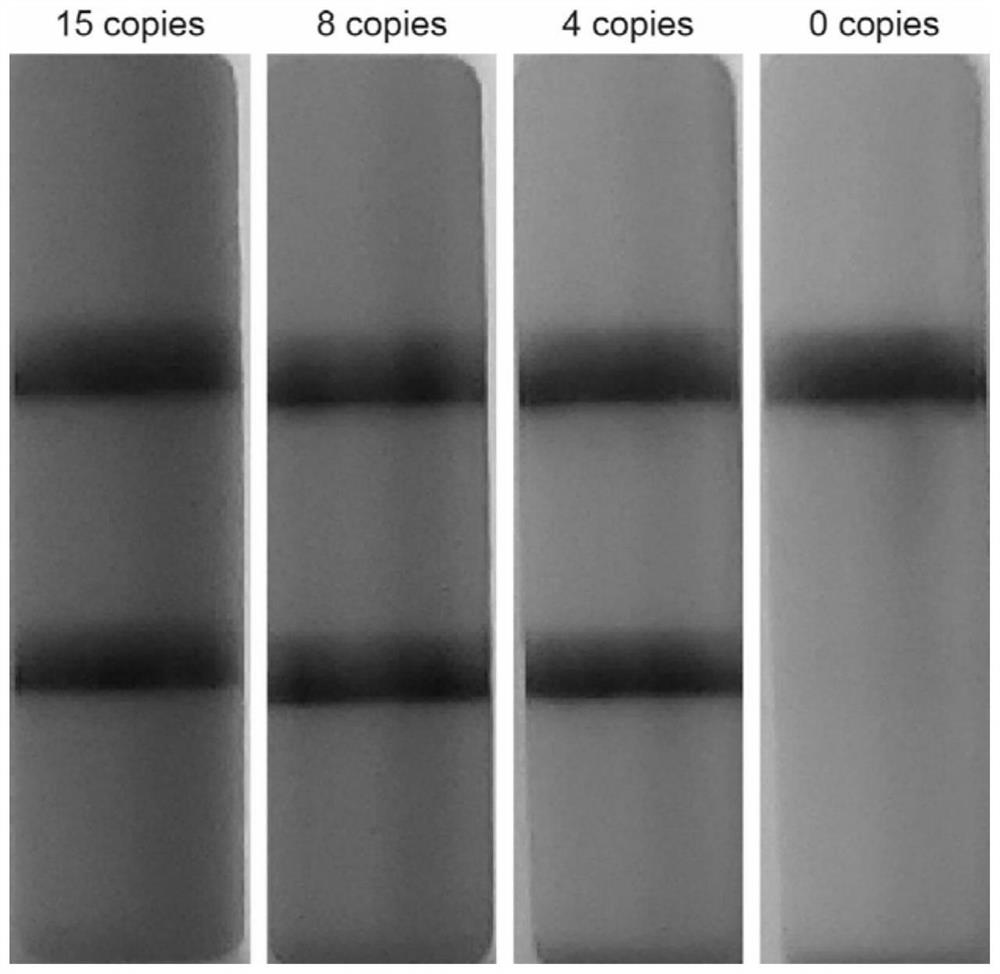
[0093] Example 2 Two-step RPA detects free viral RNA sensitivity test
[0094] The reagent preparation, experimental steps, and two-step RPA reaction system of this embodiment are the same as those in Example 1.
[0095] 1. The experimental groups are as follows:
[0096] Experimental group 1: 1 st The RNA concentration used in the RPA reaction system was 15 copies / ul. Prepared 1 st After the RPA reaction system, mix evenly by inverting, briefly centrifuge, and react at 37°C for 10 minutes. Take 10ul1 st RPA reaction product was added to 2 nd In the RPA reaction system, mix by inverting, briefly centrifuge, and react at 37°C for 10 minutes.
[0097] Experimental group two: 1 st The RNA concentration used in the RPA reaction system was 8 copies / ul. Prepared 1 st After the RPA reaction system, mix evenly by inverting, briefly centrifuge, and react at 37°C for 10 minutes. Take 10ul1st RPA reaction product and add to 2 nd In the RPA reaction system, mix by invert...
Embodiment 3
[0104] Example 3 Two-step RPA limit reaction time test
[0105] The reagent preparation, experimental steps, and two-step RPA reaction system of this embodiment are the same as those in Example 1.
[0106] 1. The experimental groups are as follows:
[0107] Experimental group 1: According to the above table, prepare 1 st For the RPA reaction system, mix evenly by inversion, centrifuge briefly, and react at 37°C for 10 minutes. Take 10ul reaction product and add to 2 nd In the RPA reaction system, mix by inverting, briefly centrifuge, and react at 37°C for 10 minutes.
[0108] Experimental group 2: Prepare 1 according to the above table st For the RPA reaction system, mix evenly by inversion, centrifuge briefly, and react at 37°C for 10 minutes. Take 10ul reaction product and add to 2 nd In the RPA reaction system, mix by inverting, briefly centrifuge, and react at 37°C for 5 minutes.
[0109] Experimental group 3: according to the above table to prepare 1 st For t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com