T7EI-like nuclease reaction buffer system, DNA mismatch detection method and application thereof
A buffer system and nuclease technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve problems such as affecting application, easy omission of SNP type differences, biased results, etc. Effects of the ability to mismatch DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] This embodiment provides a reaction buffer system for T7EI-like nuclease: including T7EI-like nuclease, Tris-HCl and Tris-Ac that provide buffering capacity, detergent Triton X-100 and Tween that enhance the dispersion of enzyme protein -20, metal ion Mg involved in substrate binding and catalysis 2+ 、Co 2+ , Mn 2+ etc., including non-specific proteins such as BSA that enhance enzyme stability, including DNA ligase that counteracts non-specific cleavage activity, etc.
[0030] The above-mentioned T7EI-like nucleases include T7EI (T7 endonuclease I) and P-SSP7EI (P-SSP7endonuclease I) and their homologous proteins; the above-mentioned proteins can be from commercial products, can be obtained by expression and purification, can be the enzyme protein itself, or It can be a fusion protein of T7EI-like protein, such as T7EI fused with maltose binding protein (MBP), and its activity needs to be calibrated before application;
[0031] The buffer capacity provider is Tris-HC...
Embodiment 2
[0038] This embodiment provides a method for DNA mismatch recognition, using the reaction buffer system of Example 1 to enhance the cleavage ability of T7EI-like nuclease, including the following steps:
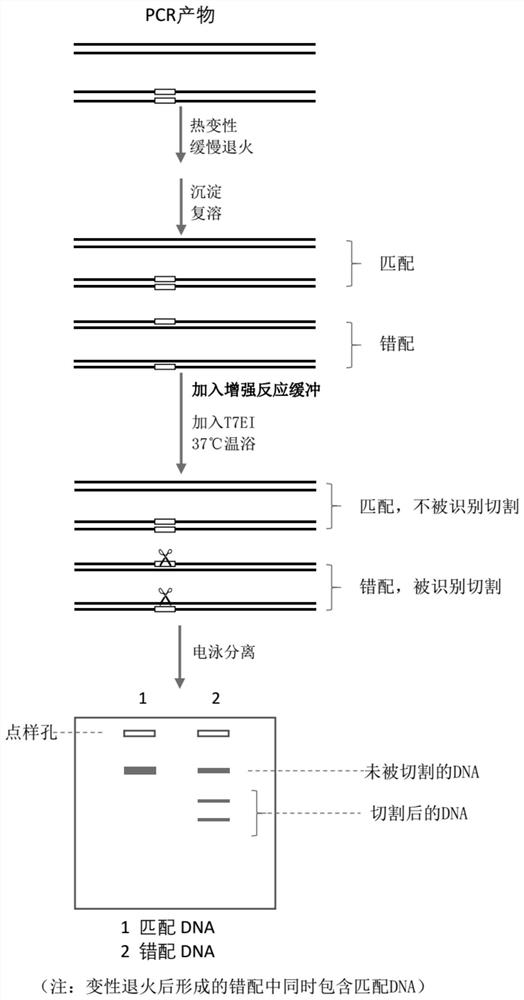
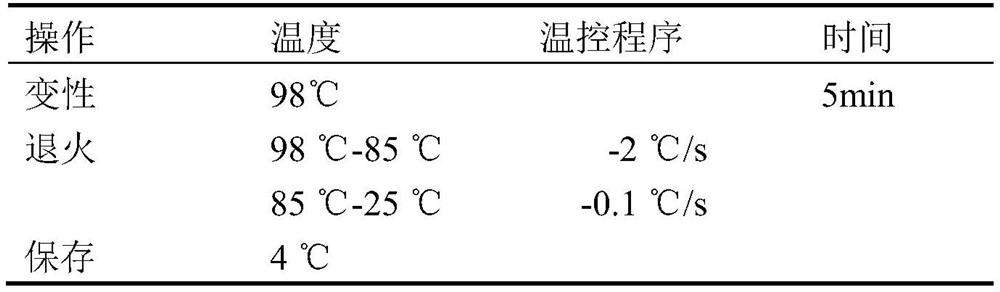
[0039] After the reaction buffer is prepared according to the method of Example 1, the sample is pretreated firstly: the PCR product or the PCR product mixture is denatured and annealed. The denaturing annealing process is shown in Table 1. Then carry out precipitation redissolution, add 1 / 10 volume of 3M potassium acetate solution (pH 5.2), add 2 times the volume of pre-cooled absolute ethanol, place on ice for 30min, centrifuge at 13000g 4°C for 10min, carefully discard the supernatant, add 1ml 70 The precipitate was washed with % ethanol, centrifuged at 13,000 g at 4°C for 10 min, the supernatant was carefully discarded, dried at room temperature, and TE buffer was added to dissolve the precipitate to obtain a mismatched substrate. If the detected mismatch type is InDel t...
Embodiment 3
[0046] This embodiment discloses the application of an enhanced reaction buffer system in various practical scenarios.
[0047] scene 1:
[0048] In the analysis of gene editing efficiency, first perform PCR amplification, denaturation annealing, precipitation and redissolution, then perform enzyme digestion according to the operation in Example 2, and then use gel electrophoresis to separate, and use gel imaging software to analyze the electrophoretic bands. The brightness is analyzed, and the efficiency of gene editing is calculated based on it. Calculated as follows:
[0049] Editing efficiency (%)=100x[1-(1-cuttable DNA ratio) 1 / 2 ]
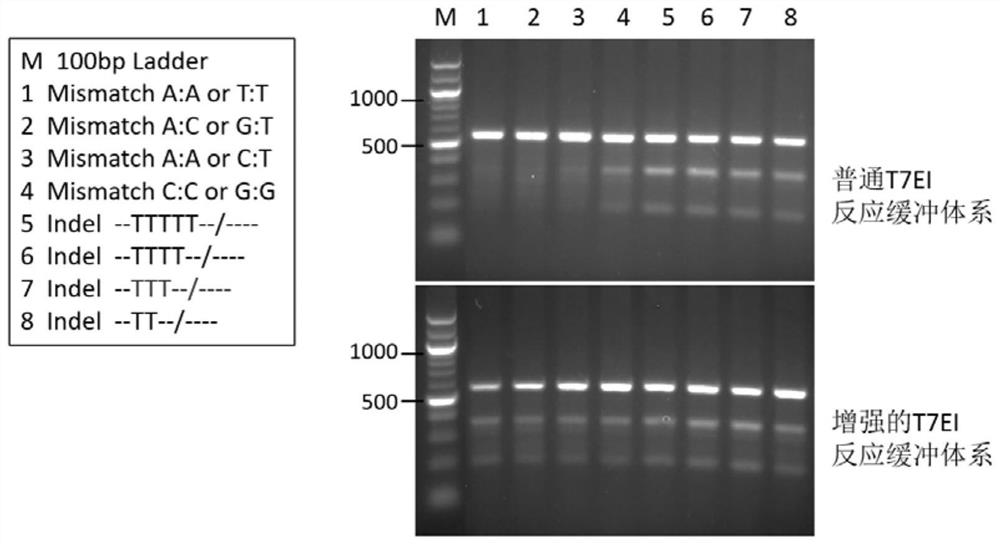
[0050] Since the T7EI-like nuclease in the reaction buffer system disclosed by the present invention can better recognize SNP type mismatches, the proportion of cleavable DNA obtained is more accurate, and the gene editing effect evaluation based on it is also more accurate.
[0051] Scenario 2:
[0052] Screening of gene-edited individu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com