Gene marker panels and their applications for the assessment of endometrial receptivity
An endometrial and receptive technology, applied in the field of biomedicine, can solve the problems of finding and infertile patients, unable to accurately and effectively assess the receptivity of the endometrium, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
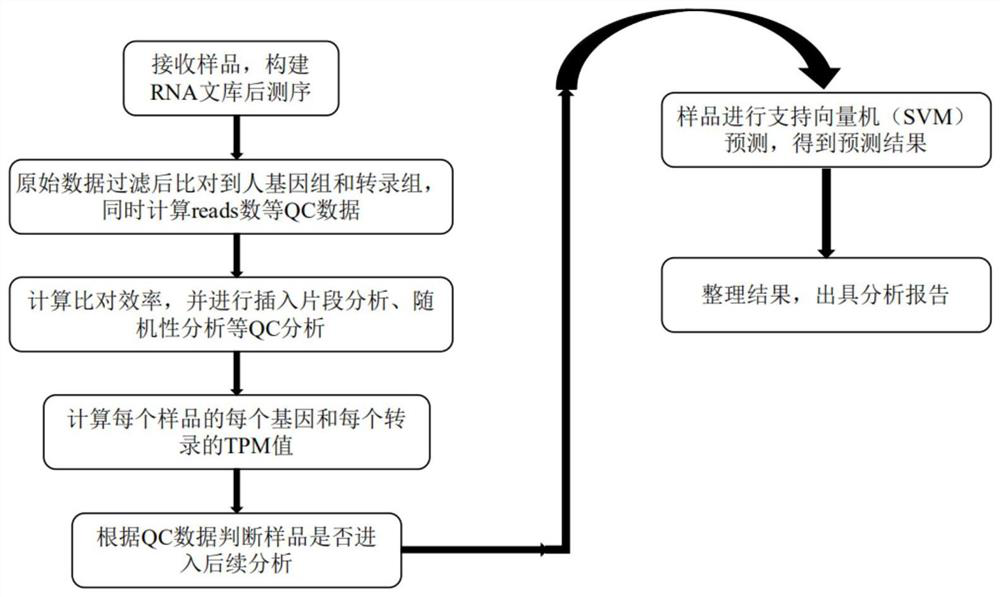
[0048] Example 1 Sample collection, transportation, quality control
[0049]1. The subject or volunteer shall be sampled by a gynecologist or a professionally qualified technician. The sample shall include not less than 1mg of endometrial tissue and not less than 4uL of uterine cavity fluid. Immediately after the biopsy was completed, the sample was added to a preservation tube containing RNA preservation solution (Qiagen RNA Later), and stored at -20°C or -80°C.
[0050] 2. Sample transportation: Use ice packs or dry ice to transport samples to designated laboratories.
[0051] 3. Sample quality control: The quality control content includes sample information, sample integrity, and the actual temperature measured when the sample is received.
[0052] 4. Specific sample collection and grouping are shown in Table 2.
[0053] Table 2 Number of samples in different periods of endometrial receptivity
[0054]
Embodiment 2
[0055] Example 2 RNA sequencing
[0056] 1. Extract total RNA from endometrial biopsy samples.
[0057] 2. Use Agilent2100 to analyze the integrity of RNA, RIN>7.0, 28S / 18S>1.2 is qualified. The RNA concentration and purity were accurately quantified using Qubit. The OD260 / OD280 was between 1.8 and 2.2, and the RNA extraction amount was greater than 2ug, which was considered qualified.
[0058] 3. Use the poly(A) purification method or the rRNA removal purification method to enrich and purify the mRNA.
[0059] 4. Use KAPA Stranded RNA-Seq Library Preparation Kit for RNA library construction.
[0060] 5. Use Agilent2100 to analyze the distribution of library fragments, and use Qubit to accurately quantify the library concentration.
[0061] 6. Use illumina Hiseq2000 on-machine sequencing.
Embodiment 3
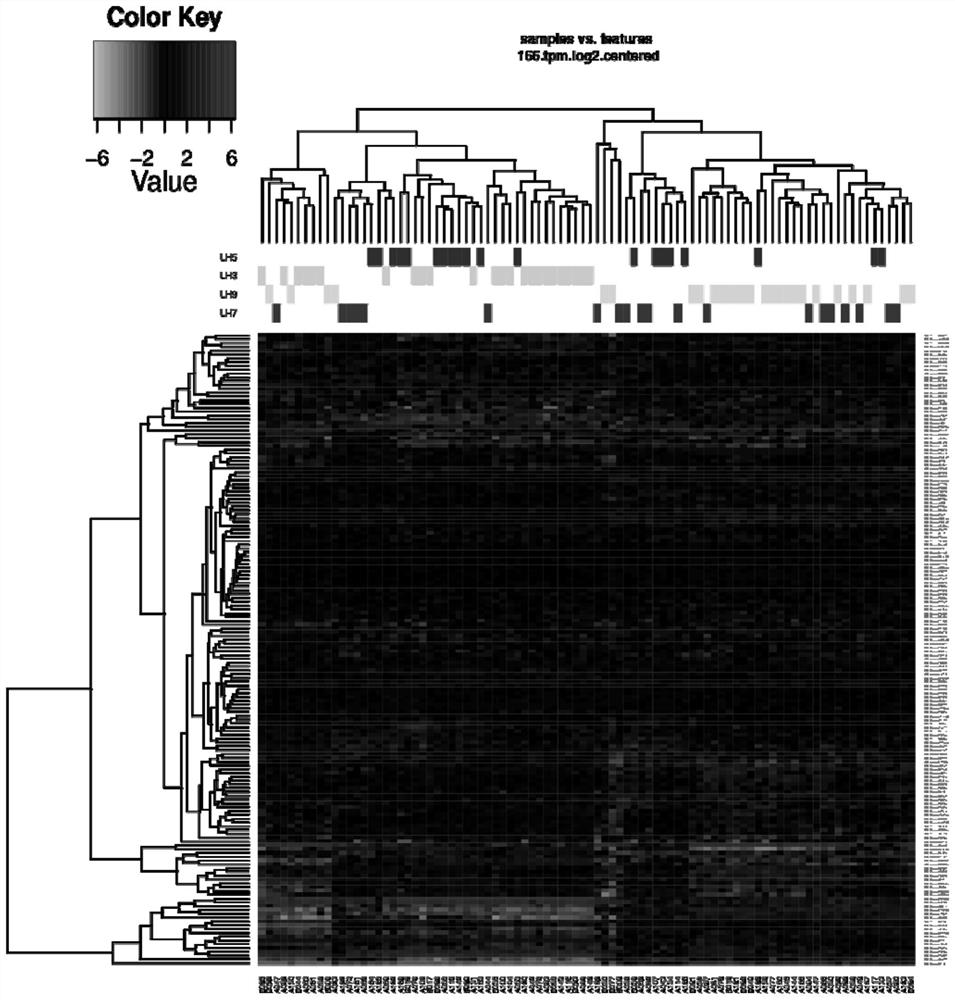
[0062] Example 3 Screening of characteristic gene markers related to endometrial receptivity assessment
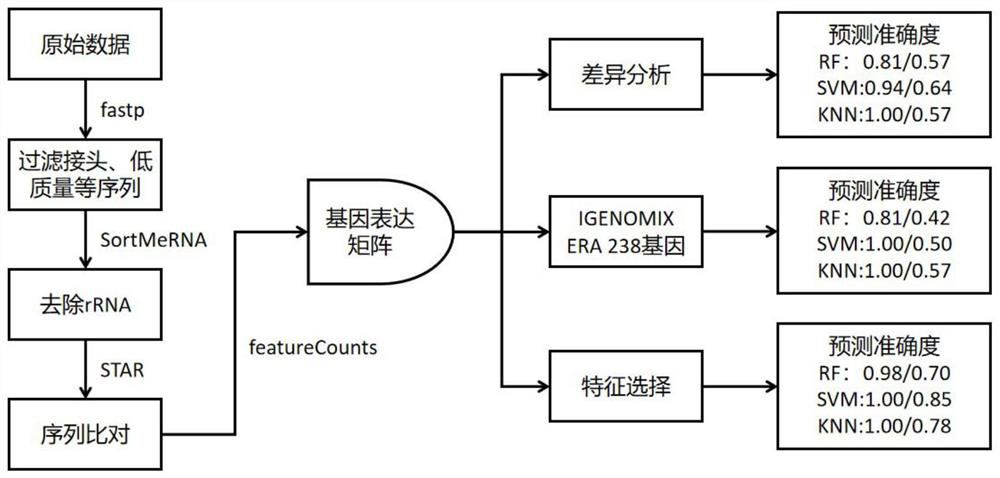
[0063] 1. Data quality control: After the original data is off the machine, the quality control of the sequencing results is carried out through the fastp software. The quality control standard is that the read length is not less than 75, and the rest of the parameters are default (the original data is called RawData, and the data after quality control is called CleanData. ).
[0064] 2. Ribosome data deletion: rRNA content can reflect the quality of library construction to a certain extent, so before comparing the reference genome, use sortmerna to compare CleanData to the ribosome library, remove the reads on the comparison, and get de - rRNA CleanData default filter parameter is 1e-5.
[0065] 3. Reference genome comparison: compare de-rRNA CleanData to the reference genome or transcriptome, calculate the comparison efficiency, and perform randomness analysis and inser...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com