Biomarker detection method and system for predicting tumor immune treatment effect
A biomarker and immunotherapy technology, applied in the field of biomarker detection to predict the effect of tumor immunotherapy, can solve problems such as mismatch repair function microsatellite instability, and achieve the effect of improving accuracy and applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
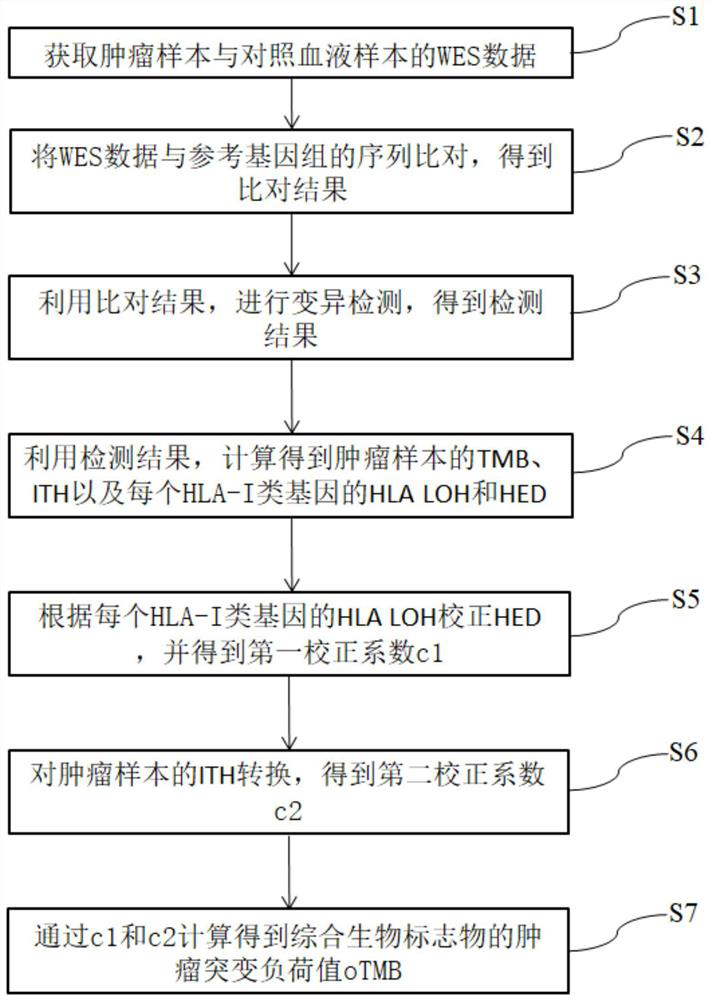
[0034] This embodiment provides a biomarker detection method for predicting the effect of tumor immunotherapy, such as figure 1 As shown, it includes the following steps:
[0035] S1. Obtain whole exome sequencing (WES) data of tumor samples and control blood samples.
[0036] Specifically, the original WES sequencing off-machine data of tumor samples and control blood samples were obtained, and the WES sequencing data were obtained after splicing and low-quality data quality control processing.
[0037] S2. Align the sequencing data with the sequence of the reference genome of the control blood sample to obtain an alignment result.
[0038] Compare the WES sequencing data obtained in step S1 with the reference genome, and remove redundancy to obtain the comparison result. In this embodiment, the reference genome is the hg19 genome, and the comparison process uses bwa (Burrow-WheelerAligne) software Then use the picard tool to remove redundancy and get the bam file of the co...
Embodiment 2
[0050] This example provides a biomarker detection method for predicting the effect of tumor immunotherapy, taking the tumor in the immunotherapy cohort of non-small cell lung cancer as an example, the specific detection method is as follows:
[0051] S1. Obtain the WES raw sequencing off-machine data of the tumor tissue samples and control blood samples of the non-small cell lung cancer immunotherapy cohort of 69 patients, and obtain the WES sequencing data after removing joints and quality control processing of low-quality data.
[0052] S2. Aligning the sequencing data of 69 cases with the sequence of the reference genome hg19 to obtain an alignment file.
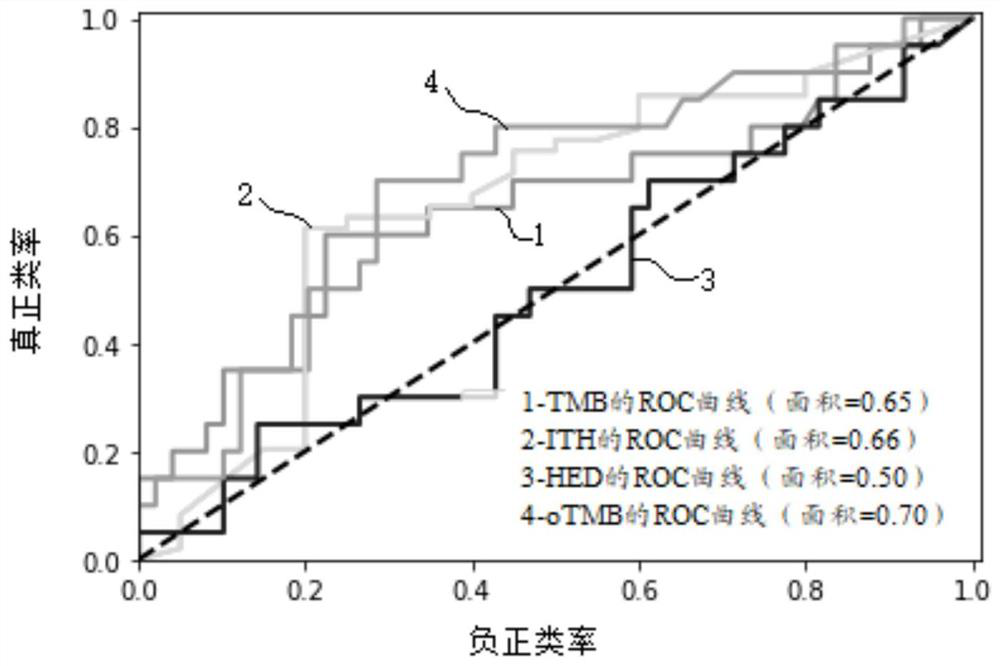
[0053] S3. Use the comparison results to detect somatic cell mutations, obtain information on somatic cell mutations, depth, mutation frequency, and somatic copy number variations of 69 patients, and obtain tumor purity, tumor ploidy, and HLA-class I genes Typing information.
[0054] S4. Using the detection results obt...
Embodiment 3
[0059] This example provides a biomarker detection method for predicting the effect of tumor immunotherapy, taking nasopharyngeal carcinoma immunotherapy cohort tumors as an example, the specific detection method is as follows:
[0060] S1. Obtain the original WES sequencing off-machine data of the tumor tissue samples and control blood samples of the nasopharyngeal carcinoma immunotherapy cohort of 61 patients, and obtain the WES sequencing data after removing joints and quality control processing of low-quality data.
[0061] S2. Aligning the sequencing data of 61 cases with the sequence of the reference genome hg19 to obtain an alignment file.
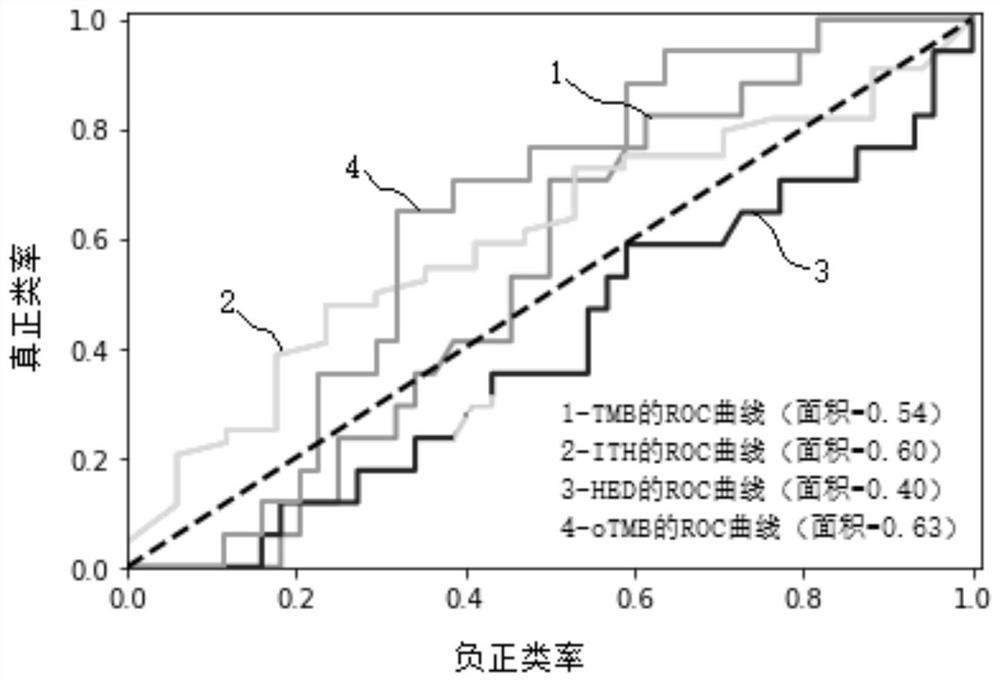
[0062] S3. Using the comparison results, somatic mutation detection was performed to obtain somatic mutation, depth, mutation frequency and somatic copy number variation information of 61 patients, as well as obtain tumor purity, tumor ploidy and HLA-class I genes Typing information.
[0063] S4. Using the detection results obtaine...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com