SNP molecular marker related to pod and seed size on peanut A06 chromosome and application thereof
A molecular marker, peanut technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. Effects of the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The character investigation of embodiment 1 natural population (250 parts of peanut materials)
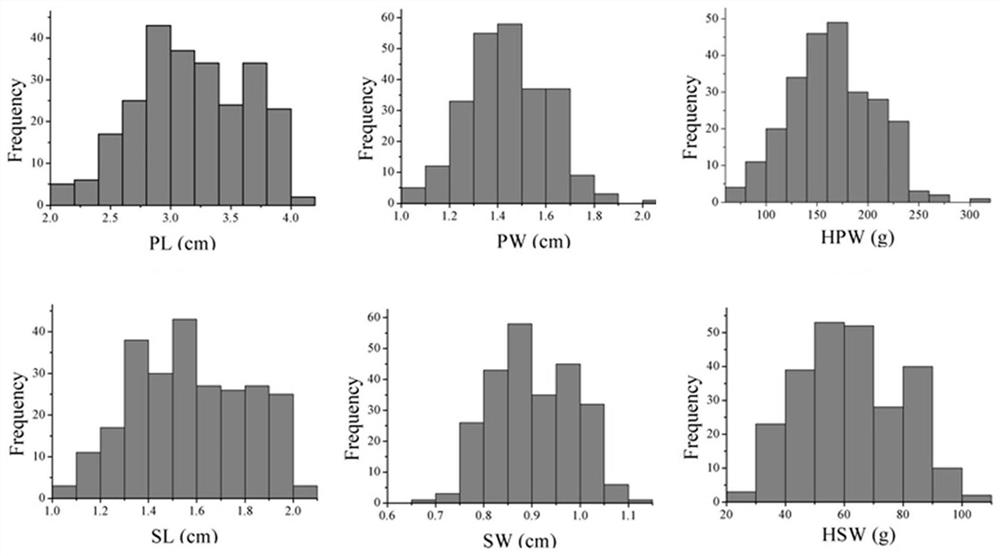
[0045] In this example, 250 peanut materials were planted in the experimental bases in Nanchong, Sichuan and Wuhan, Hubei for two consecutive years in 2015 and 2016. After ripe harvesting and drying in the sun, the characteristics of pod length, pod width, seed length, seed width, 100-fruit weight and 100-kernel weight were investigated according to the Peanut Germplasm Resources Description Specification and Data Standard (Jiang Huifang et al., 2006) (Table 1). In the four environments (the four environments are: 2015 Wuhan base (2015WH), 2015 Nanchong base (2015NC), 2016 Wuhan base (2016WH), 2016 Nanchong base (2016NC)) the average frequency is continuous distribution (see figure 1 ), indicating the quantitative genetic characteristics of these traits.
[0046] Table 1 Investigation of pod and seed size traits under four environments
[0047]
Embodiment 2
[0048] Example 2 Library Construction and Sequencing
[0049] Leaf DNA of 250 materials was extracted by CTAB method. The purity and integrity of DNA were analyzed by agarose gel electrophoresis, the purity of DNA (OD260 / 280 ratio) was detected by Nanodrop, and the DNA concentration was accurately quantified by Qubit. For qualified DNA samples, TruSeq Library Construction Kit was used to construct a GBS library using MseI, HaeIII and NlaIII restriction endonucleases, and the reagents and consumables recommended in the instructions were strictly used. DNA fragments undergo end repair, polyA tailing, sequencing adapters, purification, PCR amplification and other steps to complete the entire library preparation. After the library construction is completed, use Qubit2.0 for preliminary quantification, dilute the library to 1ng / μl, and then use Agilent 2100 to detect the inserted fragments of the library. After the inserted fragments meet the expectations, use the Q-PCR method to ...
Embodiment 3
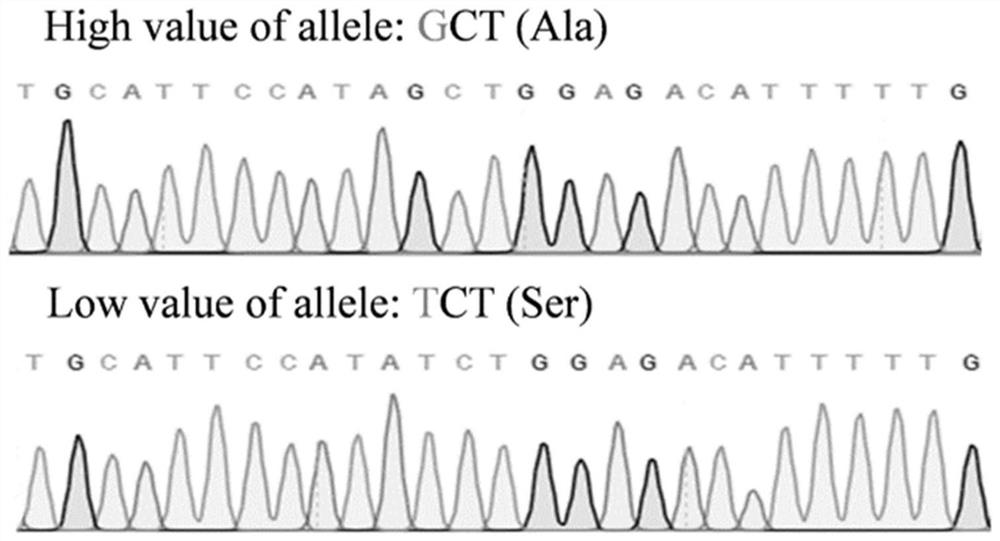
[0050] Example 3 Development of SNP markers
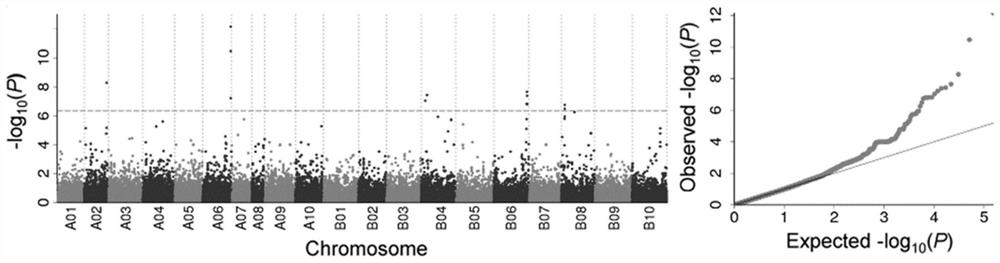
[0051] Perform quality control on the Raw data obtained after getting off the machine to obtain Clean data. The Clean data is compared to the reference genome, and the effective high-quality sequencing data is compared to the reference genome through the BWA software (with diploid A.duranensis (AA) and A.ipaensis (BB) as the reference genome, PeanutBase:http: https: / / peanutbase.org). GATK software was used to detect population SNPs. Use the Bayesian model to detect polymorphic sites in the population, and filter the detected 3,070,141 SNPs by retaining biallelic genes, miss≤0.83, and maf>0.05, and then infer by beagle, and then by GP value After filtering >0.6 and miss<0.2, 105,814 high-quality SNPs were obtained.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com